All published articles of this journal are available on ScienceDirect.
In vitro Antifungal Activity of Streptomyces Spp. against Different Barley Pathogenic Fungi
Abstract
Background:
Microorganisms and their biological products have been used as a biological control against many pathogenic. Streptomyces spp. are one of the microorganisms that can be deployed in the field as biological control of plant pathogenic fungi as an alternative to chemical pesticides.
Objective:
In this study, one hundred twenty-four isolates of Streptomyces spp. from Syrian soils were in vitro screened for their ability to inhibit the growth of various barley pathogenic fungi: Cochliobolus sativus, Pyrenophora graminea, Fusarium verticillioides, and Fusarium solani.
Method:
Streptomyces isolates which showed the highest antagonism activity for each fungus were selected.
Results:
The inhibition rate of fungal growth by the selected strains ranged between 51 and 67%. The growth inhibition of the fungus F. solani was 51% when S. malachitofuscus Str69 strain was used, compared with 67% for C. sativus by the Str74 strain. The selected Streptomyces isolates showed their production of hydrolytic enzymes that had a role in the antagonistic activity against fungi, particularly the enzymes chitinase and glucanase. The cell-free supernatants effectively inhibited the growth of the fungi, where they reduced C. sativus, P. graminea, F. verticillioides, and F. solani by 57, 68, 19, and 30%, respectively.
Conclusion:
The results of this study demonstrated that selected Streptomyces spp. isolates and their free cell crude extract could be used as a bio-control agent against the studied barley pathogenic fungi.
1. INTRODUCTION
Prevention of pathogens is critically important in agricultural production systems. Recently, the available prevention methods for conserving sustainable agriculture have been evaluated, with a focusconserving sustainable agriculture, focusing on the need to follow safe and environmentally friendly methods. Biological control of plant pathogenic fungi is a safe and good alternative to synthetic chemical pesticidessatisfying and satisfying the consumer in obtaining healthy and natural food [1, 2].
In fact, due to the ability of many microorganisms to synthesize various biological compounds structurally and functionally diverse with antagonistic activity against many organisms that cause plant diseases, it is considered active and effective agent in biological control [3]. The search for new microorganisms with more effective antifungal activity is proceeding slowly [4]. To investigate new microorganisms that are more efficient in their antagonism toward pathogenic fungi, many researches have been directed toward isolating these microorganisms from new soils and ecosystems [5, 6]. Among the microorganisms, Streptomyces spp. can be used instead of synthetic chemical fungicides and deployed as biocontrol agents.
Streptomyces are gram-positive bacteria, including about 570 species, aerobic and filamentous bacteria, capable of forming complex mycelium, and can colonize diverse environments and habitats. They are exospores-forming bacteria which enable them to spread widely [7]. Many Streptomyces spp. were widely demonstrated and used for plant growth stimulation and promotion, nutrient assimilation, biofortification, and biocontrol of plant pathogens of agriculturally important crops [8]. Cochliobolus sativus causes spot blotch, a common foliar disease, of barley, that can lead to marked reductions in both yield and the quality of the crop; therefore, it is considered an economically significant disease [9]. Pyrenophora graminea is the causal agent of barley leaf stripe disease, which often leads to a decrease in yield [10]. The genus Fusarium contains a diverse array of species responsible for damping-off, root rot and vascular wilt in a multitude of economically important plant species [11]. The present study aims to in vitro screening and test the effectiveness of some local Streptomyces isolates for their capability of being used as biocontrol agents against various barley pathogenic fungi.
2. MATERIALS AND METHODS
2.1. Fungi
Fusarium isolates: Fusarium verticillioides Fu27 and Fusarium solani Fu35, which cause head blight in cereals, were isolated from infected barley plants growing in different locations in Syria. The infected barley stems were cut into small pieces of 1-1.5cm, surfaces were sterilized with 5% sodium hypochlorite for 5min, washed in sterile distilled water twice and cultured on potato dextrose agar PDA (PDA, DIFCO, Detroit, MI. USA) medium amended with 13 mg/l kanamycin sulphate added after autoclaving and incubated for 10 days, at 23 ±1ºC in the dark to allow mycelia growth and sporulation. Species identification was based on the morphological characteristics of single-spored isolates as described by [12]. According to a study [13], the two isolates of F. verticillioides and F. solani were selected for this study. The cultures were maintained on PDA at 4ºC until needed.
Cochliobolus sativus: The strain of C. sativus pt4 that causes barley spots blotch used in the study was the most virulent of 117 isolates collected in 1998 and 2004 from naturally infected barley in different regions in Syria, as described by Arabi and Jawhar [14]. It was isolated from infected barley leaves showing spot blotch symptoms. The fungal mycelia were transferred from a stock culture into Petri dishes containing PDA with 13 mg/l kanamycin sulphate and incubated for 10 days at 22 ±1ºC in the dark to allow mycelia growth.
Pyrenophora graminea: The isolate of P. graminea Sy3, which causes barley stripe used in the study, was identified, cultivated, and maintained as described by Arabi et al. [15, 16]. It was isolated from leaf-stripe infected barley leaves from various regions in Syria and selected from among 93 isolates, according to morphological and physiological criteria (virulence). The isolates were grown separately in 9 cm Petri dishes containing PDA and incubated for 10 days at 21±1°C in the dark to facilitate mycelia growth.
2.2. Streptomyces Spp. Isolates
Various regions and soils were selected to obtain Streptomyces isolates. The soil samples were collected from different regions distributed widely from south to north-west between 33.40°N and 37.17°N and 35.40°E and 42.30°E in Syria. They were taken from 2-10 cm depth of an agriculture field and were carried to the laboratory in a sterile polythene bag. For each soil collected sample, 3 g was diluted in 100 ml of saline solution (0.85% NaCl) and allowed to stand for 15 min. Three different dilutions (1:10, 1:100 and 1:1000) were prepared using sterile saline solutions in a total volume of 10 ml. An aliquot of 0.1 ml of each dilution was plated on GYM agar (g/l): yeast extract, 4; malt extract, 10; glucose, 4; CaCO3, 2; agar, 15 [17]. To minimize the fungal and bacterial growth, nystatin 50µg/ml and polymyxin B sulphate 5.0µg/ml were added. Plates were incubated at 28 °C, and monitored after 48, 72, and 96h. Representative colonies were selected and streaked on new plates of GYM agar. The isolated Streptomyces species were preserved on GYM agar plates at 4°C until further use. Visual observation of both morphological and microscopic characteristics using light microscopy and Gram-stain properties were performed for genus confirmation. The spore suspension was prepared from the sporulated isolates after 5 days of incubation at 28°C. Spores were harvested using a sterile saline solution containing: (3 g/L Pepton, 5 g/L NaCl, 1 mL/L Tritonx100), then filtered through a layer of cotton to get rid of the mycelium hyphae and the remnants of agar. The final inoculum suspensions were adjusted at 109 spores/mL by microscopic enumeration with Neubauer improved count plate. For long storage, glycerol was added to spore suspension to make the final concentration 20% and stored at –80 °C [18].
2.3. Antagonistic activity of Streptomyces Spp. Isolates against Barley Pathogenic Fungi
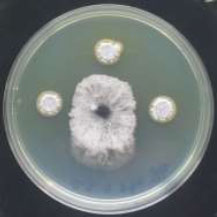
In vitro antagonism tests were performed on nutrient agar NA medium in 9cm Petri plates using a dual culture technique [19]. 5 mm diameter disc of fungi was cut from an actively growing culture by a sterile cork borer and placed onto the center of above NA, and 5ul of tested Streptomyces spore suspension was transferred from five-day-old cultures and cultivated in 2.5cm from the center of the plate, and that is on three sides of the plate, and keep the fourth side for the fungus to grow without the influence of Streptomyces as control. Secondary screening for isolates that have antagonistic efficiency was carried out, where 5ul of selected Streptomyces spore suspension adjusted at 2×109 spore/mL and pipetted in 2.5cm from the center of the plate, and the pathogenic cultivated in the center of control plate [20, 21]. The growth of the fungus was measured after a 7-day incubation period, and the antagonistic efficiency of isolates was estimated by applying the formula [22]: GI=(C-T)/C×100, where; GI=Percent inhibition, C=Radial growth of the pathogen in control, and T=Radial growth of the pathogen in treatment. Growth inhibition was categorized on a scale from 0 to 4, where 0= no growth inhibition; 1= 1 to 25%; 2= 26 to 50%; 3= 51 to 75% and 4= 76 to 100% growth inhibition [23, 24].
2.4. Identification of selected Streptomyces Strains
The primers Sm6 F (5’-GGTGGCGAAGGCGGA-3’) and Sm5 R (5’-GAACTGAGACCGGCTTTTTGA-3’) flanking a highly variable sequence region of 600 bp towards the 5’end of the 16S rDNA region were used in polymerase chain reaction PCR [25]. Genomic DNA was extracted and purified using DNA extraction DNeasy Plant Mini Kit according to the manufacturer’s recommendations (Qiagen, Cat. NO. 69104). PCR mixtures were prepared using 20ng of template DNA, 0.2μM of each primer, and a hotstar taq master mix kit (Qiagen, Cat. NO. 203446). Amplification was done under the following conditions: 10 min denaturation step at 95°C, followed by 30 amplification cycles (45 sec at 94°C, 45 sec at 65°C and 105 sec at 72°C) and an extra extension step of 10min at 72°C. PCR products were separated on a 1.5% agarose gel, to which ethidium bromide was added and photographed under UV light. Amplification products were purified using a QIAqick Gel Extraction kit (QIAGEN, Cat. No.28704) and sequenced on both strands using an ABI 310 sequencer machine. The sequences were subjected to a BLAST search against the full GenBank database available at NCBI public database using Basic Local Alignment Search Tool for Nucleotides (BLASTN) [26].
2.5. Detection of Hydrolytic Enzymes Activities by the selected Stryptomyces Isolates
The selected isolates of Stryptomyces spp. that exhibited antifungal activity against studied pathogenic fungi were cultured on a synthetic agar media to investigate their hydrolyzing enzyme production. The enzymes investigated include: chitinase, amylase, protease, glucanase, lipase, xylanase, pectinase, carboxymethylcellulase (CMCase). The detection method is based on the use of specific substrates for the studied enzymes, for example, xylan from oat spelled for xylanase, tributyrin for lipase, starch from a potato for amylase, and colloidal chitin for chitinase. The activated culture of each isolate was streaked on agar media containing a specific substrate for each enzyme activity. The presence of hydrolytic activities for some enzymes had determined by staining agar cultures with congo red solutions. Isolates that have hydrolytic enzyme activity showed clearing zones on agar media [27], and the hydrolytic efficiency of isolates was estimated 5 days after incubation at 28°C by calculating the ratio of clear zone CZ diameter to growth zone GZ (CZ/GZ) [28].
2.6. The Inhibitory Effect of Cell-free Supernatant of Streptomyces ssp. (in vitro)
A liquid culture of selected isolates was performed on nutrient broth NB medium with 7% (w/v) wet mycelium fungi and incubated at 28°C with constant shaking at 200 rpm for 7 day. The culture was removed and centrifuged at 8000×g for 10min. Cell-free supernatant was obtained by filter (0.45μm) sterilized and tested for antifungal activity [29]. Four wells (5 mm in diameter) were made in an NA plate on the side 2.5cm from the center of the plate using a sterilized cork borer, subsequently wells were filled with 200μl of filtered supernatant [21]. Five mm diameter disc of fungi, taken from the edge of an actively growing fungal colony, was inoculated in the center of NA plate [31], and compared with control plates. Plates were incubated for 5 days at 28°C. Triplicate plates were performed for each target fungal organism and radial growth inhibition of the fungus was measured.
2.7. Statistical Analysis
The statistical analysis was done using the Statistica program version 6 (Statsoft, Inc. 2003) at 5% significance level (P = 0.05). Data were subjected to the analysis of variance for determining the statistical significance of differences between means, according to the ANOVA-Tukey HSD test.
3. RESULTS AND DISCUSSION
3.1. Screening of Streptomyces Isolates for Antagonistic activity against studied Plant Pathogenic Fungi
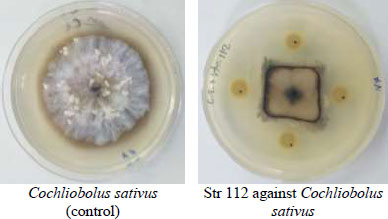
The microbial antagonist was considered the promising biocontrol strategy due to high efficiency, broad spectrum and environmental friendliness [31]. Few Actinobacteria were identified and employed due to their differences in growth conditions and antagonistic activities. Therefore, isolation and screening of highly efficient antagonistic Actinobacteria are keys to developing some biocontrol agents [32]. One hundred and twenty-four isolates of Streptomyces were tested for their efficacy in inhibiting the growth of the studied pathogenic fungi (Fig. 1). Only 25 of them showed antagonistic activity against Fusarium verticillioides and F. solani, whereas 90 of them showed antifungal efficacy against Cochliobolus sativus and Pyrenophora graminea. Only 13% of isolates showed antagonistic ability against all of the tested fungi, while 15% of them did not show any antagonism. A secondary screening was carried out for the isolates, which inhibited the growth of the fungi (Fig. 2). According to a study [22], the results were categorized on a scale from 1 to 3, where 1: 1 to 25%, 2: 26 to 50% and 3: 51 to 75%. The results showed that 4 isolates have high inhibition rate against P. graminea, 3 isolates against C. sativus, 2 isolates against F. verticillioides and 2 isolates against F. solani (Table 1). The isolates: Str74, Str18, Str72, Str69 exhibited the highest growth inhibition of C. sativus (67%), P. graminea (56%), F. verticillioides (66%) and F. solani (62%), respectively. This clearly suggested that these isolates possess antagonism against fungi in vitro, which may contribute to the production of antifungal compounds that reduced fungi's mycelial growth by forming an inhibition zone. The antagonistic activity of Streptomyces to fungal pathogens is usually related to the production of antifungal compounds and extracellular hydrolytic enzymes [33, 34]. Many species of Actinomycetes, particularly those belonging to the genus Streptomyces, are well-known as antifungal biocontrol agents that inhibit several plant pathogenic fungi [35, 36]. Several researchers have already reported similar antifungal activity of Streptomyces against fungal pathogens. The percentage of isolates that showed the ability and efficiency of inhibition varied according to the studied Streptomyces isolates and the target fungi [37-40]. For example, Table 2 shows the results of antagonisms in similar studies and their comparison with our results.

(Explanation of category 1): Percent growth inhibition was determined 7 days after incubation using Whipps’ (1987) formula. Values were categorized on a scale from 0 to 3, where 0: no growth inhibition; 1:1 to 25%; 2: 26 to 50%; 3: 51 to 75%.
| Streptomyces spp. | GI Category1 | Streptomyces spp. | GI Category1 | Streptomyces spp. | GI Category1 | Streptomyces spp. | GI Category1 | ||||||||||||
| C. Sativus | P. Graminea | F. Verticillioides | F. Solani | C. Sativus | P. Graminea | F. Verticillioides | F. Solani | C. Sativus | P. graminea | F. Verticillioides | F. Solani | C. Sativus | P. Graminea | F. Verticillioides | F. Solani | ||||
| Str 1 | 2 | 1 | 1 | 1 | Str 32 | 2 | 1 | 2 | 2 | Str 63 | 0 | 1 | 0 | 0 | Str 94 | 0 | 0 | 0 | 0 |
| Str 2 | 1 | 1 | 0 | 0 | Str 33 | 2 | 2 | 0 | 0 | Str 64 | 0 | 0 | 0 | 0 | Str 95 | 0 | 1 | 0 | 0 |
| Str 3 | 1 | 1 | 0 | 0 | Str 34 | 0 | 1 | 0 | 0 | Str 65 | 2 | 2 | 2 | 2 | Str 96 | 0 | 2 | 0 | 0 |
| Str 4 | 1 | 0 | 1 | 1 | Str 35 | 0 | 0 | 0 | 0 | Str 66 | 0 | 0 | 0 | 0 | Str 97 | 2 | 0 | 0 | 0 |
| Str 5 | 1 | 0 | 2 | 2 | Str 36 | 1 | 1 | 0 | 0 | Str 67 | 3 | 2 | 2 | 2 | Str 98 | 2 | 1 | 0 | 0 |
| Str 6 | 1 | 0 | 1 | 1 | Str 37 | 0 | 0 | 0 | 0 | Str 68 | 0 | 1 | 0 | 0 | Str 99 | 2 | 1 | 0 | 0 |
| Str 7 | 1 | 0 | 1 | 1 | Str 38 | 0 | 2 | 0 | 0 | Str 69 | 2 | 0 | 3 | 3 | Str 100 | 2 | 1 | 0 | 0 |
| Str 8 | 1 | 1 | 1 | 1 | Str 39 | 0 | 2 | 0 | 0 | Str 70 | 0 | 1 | 0 | 0 | Str 101 | 1 | 2 | 0 | 0 |
| Str 9 | 1 | 1 | 1 | 1 | Str 40 | 0 | 2 | 0 | 0 | Str 71 | 0 | 0 | 0 | 0 | Str 102 | 0 | 0 | 0 | 0 |
| Str 10 | 1 | 1 | 1 | 1 | Str 41 | 1 | 1 | 0 | 0 | Str 72 | 2 | 1 | 3 | 3 | Str 103 | 2 | 1 | 0 | 0 |
| Str 11 | 1 | 1 | 1 | 1 | Str 42 | 1 | 1 | 0 | 0 | Str 73 | 3 | 1 | 0 | 0 | Str 104 | 0 | 1 | 0 | 0 |
| Str 12 | 1 | 1 | 0 | 0 | Str 43 | 1 | 1 | 0 | 0 | Str 74 | 3 | 0 | 2 | 2 | Str 105 | 0 | 2 | 0 | 0 |
| Str 13 | 1 | 1 | 2 | 2 | Str 44 | 1 | 1 | 0 | 0 | Str 75 | 0 | 0 | 0 | 0 | Str 106 | 2 | 1 | 0 | 0 |
| Str 14 | 1 | 1 | 1 | 1 | Str 45 | 0 | 0 | 0 | 0 | Str 76 | 0 | 1 | 0 | 0 | Str 107 | 2 | 0 | 0 | 0 |
| Str 15 | 1 | 1 | 1 | 1 | Str 46 | 2 | 0 | 0 | 0 | Str 77 | 0 | 0 | 0 | 0 | Str 108 | 0 | 0 | 0 | 0 |
| Str 16 | 2 | 1 | 1 | 1 | Str 47 | 1 | 2 | 2 | 2 | Str 78 | 0 | 0 | 0 | 0 | Str 109 | 2 | 1 | 0 | 0 |
| Str 17 | 0 | 3 | 2 | 2 | Str 48 | 2 | 1 | 0 | 0 | Str 79 | 2 | 1 | 0 | 0 | Str 110 | 2 | 2 | 0 | 0 |
| Str 18 | 0 | 3 | 0 | 0 | Str 49 | 2 | 2 | 0 | 0 | Str 80 | 2 | 2 | 2 | 2 | Str 111 | 1 | 1 | 0 | 0 |
| Str 19 | 1 | 1 | 0 | 0 | Str 50 | 2 | 2 | 0 | 0 | Str 81 | 0 | 0 | 0 | 0 | Str 112 | 3 | 0 | 0 | 0 |
| Str 20 | 2 | 1 | 0 | 0 | Str 51 | 2 | 0 | 0 | 0 | Str 82 | 2 | 1 | 0 | 0 | Str 113 | 1 | 1 | 0 | 0 |
| Str 21 | 2 | 2 | 0 | 0 | Str 52 | 0 | 0 | 0 | 0 | Str 83 | 1 | 1 | 0 | 0 | Str 114 | 2 | 2 | 0 | 0 |
| Str 22 | 1 | 1 | 0 | 0 | Str 53 | 0 | 0 | 0 | 0 | Str 84 | 1 | 1 | 0 | 0 | Str 115 | 1 | 1 | 0 | 0 |
| Str 23 | 1 | 1 | 0 | 0 | Str 54 | 0 | 0 | 0 | 0 | Str 85 | 0 | 2 | 0 | 0 | Str 116 | 1 | 2 | 0 | 0 |
| Str 24 | 1 | 1 | 0 | 0 | Str 55 | 2 | 2 | 0 | 0 | Str 86 | 1 | 2 | 0 | 0 | Str 117 | 3 | 2 | 2 | 2 |
| Str 25 | 2 | 2 | 0 | 0 | Str 56 | 0 | 0 | 0 | 0 | Str 87 | 2 | 2 | 0 | 0 | Str 118 | 2 | 0 | 0 | 0 |
| Str 26 | 1 | 1 | 0 | 0 | Str 57 | 1 | 0 | 2 | 2 | Str 88 | 2 | 2 | 0 | 0 | Str 119 | 2 | 2 | 0 | 0 |
| Str 27 | 0 | 1 | 0 | 0 | Str 58 | 0 | 0 | 0 | 0 | Str 89 | 0 | 1 | 0 | 0 | Str 120 | 1 | 1 | 0 | 0 |
| Str 28 | 2 | 1 | 0 | 0 | Str 59 | 1 | 0 | 0 | 0 | Str 90 | 0 | 0 | 0 | 0 | Str 121 | 2 | 2 | 2 | 2 |
| Str 29 | 1 | 1 | 0 | 0 | Str 60 | 0 | 1 | 0 | 0 | Str 91 | 2 | 3 | 0 | 0 | Str 122 | 2 | 2 | 0 | 0 |
| Str 30 | 1 | 1 | 0 | 0 | Str 61 | 1 | 0 | 0 | 0 | Str 92 | 0 | 1 | 0 | 0 | Str 123 | 2 | 2 | 0 | 0 |
| Str 31 | 1 | 1 | 0 | 0 | Str 62 | 2 | 1 | 0 | 0 | Str 93 | 1 | 0 | 0 | 0 | Str 124 | 3 | 2 | 0 | 0 |
| References | The Highest Rate of Growth Inhibition | Percentage or Number of Isolates that have Antifungal Activity among the Studied Isolates | Fungi | Streptomyces Strains |
| [42] | 33.7% | 2 isolates | C. sativus | Streptomyces sp. R18(6) |
| [42] | 44.6% | 2 isolates | Streptomyces sp. 6(4) | |
| Our work | 67% | 90% | Streptomyces sp. Str74 | |
| - | - | - | P. graminea | No published results were found |
| Our work | 56% | 90% | Streptomyces sp. Str18 | |
| [41] | 34 mm≤ | 20% | F. verticillioides | S. spororaveus RDS28 |
| [43] | 33% | 4 strains | Streptomyces sp. ST08 | |
| Our work | 66% | 25% | Streptomyces sp. Str72 | |
| [41] | 36 mm≤ | 4% | F. solani | S. spororaveus RDS28 |
| [43] | 25% | 4 strains | Streptomyces sp. ST02 | |
| Our work | 62% | 25% | Streptomyces sp. Str69 |
| Enzymatic Hydrolysis Efficiency (CZ/GZ) | Streptomyces Isolates | |||||||
| Amylase | Chitinase | Lipase | Protease | Xylanase | Pectinase | Glucanase | CMCase | |
| 2.8 | 1.1 | 1.2 | 1.9 | 1.1 | 1.2 | 1.3 | 1.7 | Str18 |
| 1.3 | 1.3 | 1.4 | 2.7 | 2.8 | 3.9 | 2.8 | 2.4 | Str69 |
| 1.5 | 1.2 | 1.3 | 2.8 | 3.1 | 3.8 | 2.5 | 2.6 | Str72 |
| 1.2 | 1.2 | 1.3 | 2.6 | 2.9 | 4.3 | 2.4 | 2.3 | Str74 |
| 2.6 | 2.6 | 1.2 | 2.1 | 1.1 | 1.1 | 1.2 | 1.8 | Str91 |
| 1.4 | 1.9 | 1.4 | 0.0 | 1.5 | 4.3 | 4.6 | 2.4 | Str98 |
| 2.3 | 1.2 | 1.2 | 2.9 | 2.8 | 3.7 | 3.9 | 2.9 | Str112 |
| Average Growth Diameter (mm)* for Each Fungus | Treatment | Fungi |
| 52.66 ± 1.15 A | Control | Cochliobolus Sativus |
| 32.66 ± 1.53 B | Str. 112 | |
| 22.67 ± 1.52 C | Str. 74 | |
| 59.00 ± 1.00 A | Control | Pyrenophora graminea |
| 20.50 ± 0.50 B | Str. 91 | |
| 19.00 ± 1.00 B | Str. 18 | |
| 64.00 ± 1.00 A | Control | Fusarium Verticillioides |
| 54.00 ± 1.00 B | Str. 69 | |
| 52.00 ± 2.00 B | Str.72 | |
| 84.67 ± 0.67 A | Control | Fusarium Solani |
| 73.33 ± 1.53 B | Str. 69 | |
| 59.17 ± 0.76 C | Str. 72 |
3.2. Streptomyces Strains Identification
Identification of selected isolates was performed using 16S DNA gene sequences. PCR amplification with specific primers Sm yielded single DNA fragments of ~ 600 bp, present in all Streptomyces sp. isolates (Fig. 3). The nucleotide BLAST similarity search analysis, based on 16S DNA gene sequence, revealed that these isolates belong to the Streptomyces genus. The closest phylogenetic neighbors according to the 16S DNA gene sequence data were S. malachitofuscus, with 99% of homology (Accession number MZ027071) for Str69, S. neyagawaensis, with 99% of homology (Accession number ON076881) for Str18, S. nodosus with 97% of homology (Accession number ON568310) for Str72, and S. Streptomyces albidoflavus with 99% of homology (Accession number ON076879) for Str74. Several studies have exposed the possibility of using Streptomyces species to protect cereal crops from diseases, especially those caused by fungal [43].
3.3. Detection of Extracellular Hydrolytic Enzymes Activities
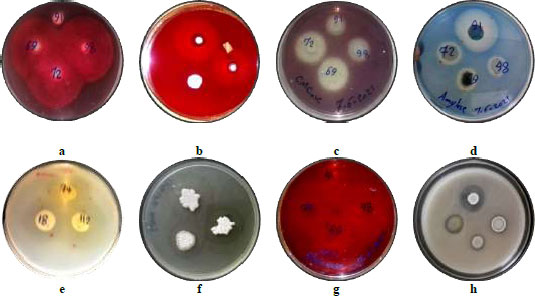
Streptomyces sp. in the phyla Actinobacteria are broadly identified for the commercial production level of secondary metabolites and enzymes in a variety of applications viz. medicine, agriculture and environment sectors. The results of the enzymes activities of the selected isolates included chitinase, amylase, protease, glucanase, lipase, xylanase, pectinase, carboxymethyl cellulase (CMCase) showed a different efficiency of the tested hydrolytic enzymes (Table 3). Where these isolates were able to hydrolyze substrates and form clear zones around the growth zones (Fig. 4), and played an important role in their antifungal activity. The inhibition of fungi by Streptomyces may be related to the production of chitinase [44]. Several Streptomyces species produce enzymes that degrade chitin, which is the major component of most fungal cell walls. Therefore, chitinases are used as potential biocontrol agents for many fungal pathogens through their chitin degradation activity. In a study by Baharlouei et al. [45], they reported 18 Streptomyces strains out of 110 isolates had strong chitinase activity. Since the fungal cell wall contains a matrix of proteins, proteases play a significant role in the degradation of the wall [46]. Glucans are the most abundant polysaccharides in the cell walls of fungi, therefore, glucanase has received great attention in recent years regarding their potential antifungal activities. Microorganisms that produce chitinase, glucanase and other hydrolytic enzymes are biological control agents that can control fungi that cause plant diseases [47, 48].
3.4. In vitro Inhibitory Effect of Cell Free Supernatant of Streptomyces sp.
Current study results showed that the cell-free supernatant of Streptomyces sp. strains was sufficient to inhibit fungi mycelium growth (Table 4). Cell-free supernatants of Str112 and Str74 reduced the growth of C. sativus by 38 and 57%, respectively; Str91 and Str18 inhibited P. graminea by 65 and 68%, respectively; while F. verticillioides and F. solani were more resistant as treatment with cell-free liquid of Str69 and Str72 reduced the growth just for 19 to 30% compared to the control. It was noted that the antifungal activity of the selected strains was higher than that of their cell-free extracts, except for the Str18 strain, where cell-free extract had higher efficiency. This may be explained by observing the strains were more efficient in producing the hydrolytic enzymes on agar media than the Str18 strain (Table 3), and by producing another antifungal compound in Str18 liquid culture. Biological controls using microorganisms are an appropriate substitute for such negative impacts on the environment and disease management. Such biological control measures are well adapted to the soil ecology and determinedly exert effective antagonistic activity against soil-dwelling plant pathogens [49]. Actinobacteria represent an extensive array of important and renowned resources for bioactive metabolites, among the members of Streptomyces and it contributed more than 60% of the antimicrobials compounds to date [33, 50]. Recently, the antimicrobial compounds detected in Streptomyces sp.YYS-7 in the GC-MS fractions contained the highest levels of phenolic substance [32]. Also, hydrocarbon compounds were confirmed to possess an antagonistic potential against a wide range of pathogens [51]. Additionally, the accumulated evidence showed that methyl stearate had a high antibacterial and antioxidant ability [52]. Thus, we proposed that these compounds could be the key contributor to the antimicrobial activity of Streptomyces spp. isolates. Especially, some unknown function metabolites will be the subject of future investigation. It suggested that Streptomyces spp. can be a potential antifungal agent for the inhibition of plant fungal diseases.
CONCLUSION
The results of this study demonstrated the ability of the selected Streptomyces spp. isolates to inhibit the growth of plant pathogenic fungi that were in vitro studied. This antifungal activity of the above-mentioned Streptomyces strains was associated with the production of hydrolytic enzymes, especially chitinase and glucanase, that have a possible role in their antagonistic ability against fungi and may be associated with the production of other antifungal compounds. Also, cell-free crude extract of Streptomyces liquid culture showed ability to inhibit the growth of the fungi. The results recorded an inhibitory role of Streptomyces spp. for the pathogenic fungus Pyrenophora graminea, which was not mentioned previously. We will investigate the optimization of enzyme production from the selected strains and test their antagonism against fungi in vivo.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We would like to thank the Director General of AECS and the Head of the Molecular Biology and Biotechnology Department for their much-appreciated help throughout the period of this research. We are also grateful to Miss Doaa Abdullah for her help carrying out some experiments.


