All published articles of this journal are available on ScienceDirect.
Use of a Consortium of Agronomically Important Microorganisms for Growing Soybean (Glycine max (L.) Merr.)
Abstract
Aims:
This study aimed to create a consortium of agronomically important microorganisms based on local strains of rhizobia and phosphate-solubilizing bacteria that could increase the nitrogen and phosphorus nutrition of soybean, thereby increasing its productivity in Kazakhstan.
Background:
The use of agronomically important microorganisms that simultaneously possess several useful properties for growing plants is a priority for the sustainable development of organic agriculture.
Objective:
The objectives of the study were to isolate and functionally characterize rhizobia from soybean nodules (Glycine max (L.) Merr.) and phosphate-solubilizing bacteria from rhizosphere soybean.
Methods:
In this study, local rhizobia and phosphate-solubilizing bacteria adapted to the soil and climatic conditions of Kazakhstan were isolated from the nodules and rhizosphere of soybean. The nitrogenase activity of rhizobia was determined by the acetylene reduction assay. The biocompatibility of consortium strains was determined by the perpendicular streak technique. The plant growth-promoting activity, nitrogenase, phosphate-solubilizing activity, and nodulation of isolated bacteria were studied, and the four most active strains were selected. Identification of these strains was carried out by sequencing the 16S rRNA. Consortia of agronomically important microorganisms were created based on active strains of rhizobia and phosphate-solubilizing bacteria.
Results:
The sequencing of 16S rRNA of the selected strains showed that rhizobia belonged to the genus Rhizobium and the phosphate-solubilizing to the genera Pseudomonas and Enterobacter. The results showed that seeds inoculation by consortia had a highly stimulating effect on soybean plants' growth and significantly increased the stem height (1.8-2.0 times), root length (2.3-2.7 times), and the number of nodules (2.7-3.2 times) compared to the control without inoculation. Besides, these consortia induced a significant increase in the number of nodules on soybean roots and their nitrogen fixation, an increase in phosphorus absorption, and an increase in protein in soybean plants compared to the control. According to these results, consortium No. 21 was selected as the most effective one. The consortium included strains of rhizobia Rhizobium lupini RH-7 and phosphate-solubilizing bacteria Pseudomonas koreensis FT-4.
Conclusion:
A consortium of agronomically important microorganisms based on local strains of bacteria adapted to the soil and climatic conditions and not competing with microbes of the rhizosphere was created in our study. The use of a consortium based on local strains will help avoid competition with the indigenous populations of rhizosphere bacteria, and it can be used to grow an economically important crop, such as soybean.
1. INTRODUCTION
Legumes are one of the most important food crops, with rising demands worldwide due to their high protein content. Legumes are characterized by high protein content and serve as the main source of protein in countries where meat and dairy products are not available for several reasons [1]. Soybean occupies a leading place in importance among the different legumes. Soybean is cultivated in more than 60 countries, and its production is increasing annually worldwide. According to the FAO (Food and Agriculture Organization of the United Nations), soybean was cultivated in 2018 on 121.53 million hectares, accounting for a total production of 334.89 million tons. It is projected that soybean production will almost double by 2050 [2]. Globally, soybean ranks fourth after wheat, rice, and corn and is regarded as the highest quality and cheapest solution to the problem of protein deficiency [3]. The high content of high-grade protein in soybean (up to 48%), high-quality oil (up to 25%), and vitamin B (3 times more than in cow's milk) determine its widespread use [4]. At the same time, soybean is rich in fiber; its use in human nutrition reduces cholesterol levels, controls blood sugar levels, and helps fight obesity [5]. Although soy is used in human nutrition, the majority (77%) of soy in the world is fed to animals for meat and milk production. Of these, 7% is fed in the form of beans, and the rest is first processed and only then used for livestock feed [6]. Brazil, the USA, and Argentina are the leading countries in soybean production, which jointly account for 81% of the world’s production [7, 8].
Although soybean is a significant crop as a food product, due to their cultivation, a reduction has been reported in forest areas, especially in Latin America. According to a forecast, soybean production in Brazil in the next decade will grow by about 33% and reach 151.9 million tons. By 2029, Brazil and Argentina will grow soybeans on more than 95 million ha, leading to increased areas for crops through the clearing of tropical forests. Over the past 10 years, the area under soybean crops has already increased by 67% [9]. This trend is observed all over the world. In the period 1990-2020 alone, the area of forests in the world in absolute terms decreased by 178 million ha, and soybean cultivation is the main cause of deforestation [10]. In this regard, increasing the productivity of soybeans without increasing the area of their sowing is very relevant.
For Kazakhstan, soybean is an extremely popular crop, as it reduces protein deficiency in people and solves the problem of a shortage of animal feed [11]. However, its soybean yields are low compared to other countries. Thus, the average productivity of soybean in Brazil and the USA is 3.3 t/ha, and in Canada, it is 2.6 t/ ha, while in Kazakhstan, on average, it does not exceed 1.1-1.2 t/ha. At the same time, soybean grain is characterized by low-quality indicators [12].
To increase the productivity of soybean, mineral nitrogen and phosphorus fertilizers are used. The use of mineral fertilizers allows for achieving high yields of agricultural products, but leads to negative consequences, including soil acidification, heavy metal pollution, soil compaction, and changes in soil microbiome bacterial composition and microbial metabolic activity, resulting in a reduction of beneficial bacteria [13].
At the same time, nitrogen and phosphorus are the main elements of nutrition for plants, and it is impossible to grow crops without them. Thus, nitrogen is the limiting factor for crop productivity; the lack of nitrogen causes a significant delay in plant growth, slows down its flowering and bean setting, and causes a general weakening of plants [14]. To improve the nitrogen nutrition of soybean, both mineral and biological fertilizers are used, but the emphasis is on the use of biological preparations (bio inoculants) based on nodule symbiotic nitrogen-fixing bacteria, such as rhizobia. A large number of preparations for soybean based on rhizobia are produced in the world [15-17]. The main producers are the USA with 20 million hectares/doses annually, Brazil with 4-6 million hectares/doses, and India with 2-4 million hectares/doses [18]. In Kazakhstan, the rhizobia preparations used are mainly of imported origin. The use of imported biopreparations is often not effective due to the poor survival of rhizobia on the roots of local soybean varieties and their lack of adaptation to the soil and climatic conditions of Kazakhstan [19, 20].
Another important element of plant nutrition is phosphorus, which accelerates the ripening process and increases the stress resistance of crops to abiotic (salinity, drought, cold, etc.) and biotic factors (phytopathogens, insect pests, etc.) [21]. Agriculture around the world is directly dependent on phosphorus as an essential plant nutrient. Most agricultural soils contain high reserves of phosphorus. However, the amount of soluble phosphorus available for plants in the soil is very small (0.4-1.2 g/kg) [22]. To obtain high yields of crops, chemical fertilizers are used annually. According to the FAO, more than 201.5 million tons of chemical fertilizers were used in agriculture in 2019, of which 1.1 million tons of phosphate fertilizers were used in the EU [23, 24]. Phosphate in the soil is usually accumulated through the regular application of phosphate fertilizers. However, more than 80% of phosphate fertilizers applied to the soil become inaccessible to plants due to sorption or precipitation [25].
Due to climatic conditions, soybean cultivation in Kazakhstan is limited to one area, namely the Almaty region. The soils of this region are characterized by an extremely low content of mobile phosphorus (10-15 mg/kg) [26], and of this amount, only 0.01% of phosphorus is available to plants, which is due to its poor solubility and fixation in the soil [27, 28]. To obtain high yields, up to 100-120 kg/ha of phosphorus fertilizers are applied to the soil annually, of which only 8-10% is available for plants [29]. However, excessive and systematic use of phosphorus fertilizers leads to the contamination of water and soil and other negative consequences [30].
An alternative to the use of mineral phosphorus fertilizers is the use of phosphate-solubilizing microorganisms. As a result of the use of these microorganisms, up to 20-30% of phosphate forms that are difficult to be dissolved are converted into plant-accessible forms [31]. Currently, biopreparations based on phosphate-solubilizing bacteria are produced worldwide, mostly in China, Egypt, and India [32-34].
The combined use of rhizobia and phosphate-solubilizing bacteria is the most attractive way to increase soybean productivity, and the development and use of a consortium, which allows reducing the amount of nitrogen and phosphorus fertilizers, will be in great demand among farmers. The scientific novelty of our study lies in the fact that new local strains of rhizobia and phosphate-mobilizing bacteria adapted to the soil and climatic conditions of Kazakhstan are isolated and studied. In addition, the isolated strains do not enter into antagonistic relationships with soil microflora as they are natural representatives of soils. Based on local strains, a new consortium of complex effects is created, in which phosphate-solubilizing bacteria convert phosphorus compounds that are difficult to dissolve into forms accessible to plants and improve the phosphorus nutrition of plants, while rhizobia supply plants with organic nitrogen. The high practical significance of the study lies in the fact that the application of the created consortium significantly reduces the doses of mineral fertilizers or even allows completely abandoning them. The use of the consortium is also more effective than the use of expensive imported biological products since local strains adapted to the soil and climatic conditions of Kazakhstan can successfully form nodules on the roots of soybean plants.
The purpose of this study was to create a consortium of agronomically important microorganisms based on local strains of rhizobia and phosphate-mobilizing bacteria to increase the nitrogen and phosphorus nutrition of soybean and its productivity in Kazakhstan. Biopreparations based on a consortium of indigenous rhizobia and phosphate-solubilizing bacteria are more promising than the use of imported ones since the former is adapted to the local soil and climatic conditions. To achieve the goal, (1) the biological activity (plant growth promotion (PGP) activity, nitrogen-fixing, phosphate-solubilizing, nodulating ability) of rhizobia and phosphate-solubilizing bacteria isolated from soybean cultivars grown in Kazakhstan was studied, (2) molecular markers for their identification were employed, and (3) the most biocompatible strains were selected (4) to create a consortium of agronomically important microorganisms. The effects of seed inoculation with these consortia were then investigated on different varieties of soybean.
2. MATERIALS AND METHODS
2.1. Materials Used in the Study
Nodules on soybean roots of the Eureka variety (Glycine max (L.) Merr.) and phosphate-mobilizing bacteria from the rhizosphere soil of this variety were collected from soybean plants grown in the fields of the Almaty region of Kazakhstan in 2019-2021. Soybean plants belonged to different varieties, namely Eureka, Zhansaya, and Perizat, and their seeds were provided by the Kazakh Research Institute of Agriculture and Plant Growing LLC, Almalybak village, Karasai district, Almaty region, Kazakhstan (department of fodder, oil crops, and corn). The Eureka soybean variety is medium-ripened, drought-resistant, and recommended for cultivation in the Almaty and Zhambyl regions of Kazakhstan. The variety is widely used by farmers. The Zhansaya soybean variety belongs to medium-ripened varieties. It does not lodge, does not crack, its grain does not shed, and it is approved for use in the Almaty region. A new promising soybean variety Perizat belongs to the group of medium-ripened, which does not lodge, and is approved for use in the Almaty region.
2.2. Isolation of Rhizobia
Healthy and large plants with a well-developed root system and a high number of nodules on the roots of Eurika soybeans were selected for the isolation of nodules. The roots were thoroughly washed under running water. Large, pink nodules were separated from the root with tweezers and transferred to a Petri dish, where they were cut into pieces with a scalpel. For isolation of rhizobia, a bean agar medium of the following composition was used (g/l): K2HPO4: 1.0; MgSO4: 0.3; sucrose: 10.0; a decoction of 100 g of peas; agar: 20.0; pH of 6.8-7.0; table water: 1,000 ml. A small amount of nodule content with a bacteriological loop was taken, transferred to 100 µl of sterile water, and then placed on the surface of bean agar medium in a Petri dish and smeared with a spatula. With the same spatula, sequential seeding was done for another 3 dishes to obtain separate colonies [35]. The seeded dishes were incubated in a BOD incubator at 28oC. Fast-growing rhizobia appeared after 3-4 days, and the slow-growing ones after 7-9 days. The appearance of colonies within the first-second day indicated contamination of the cultures. The purity of rhizobia cultures was checked visually and under a light microscope with access to a computer (Premiere MAX-200, China) with preparations of live and fixed colored cells.
2.3. Nodulating Ability of Rhizobia
To study the nodulating ability of rhizobia, the bacteria were grown on a liquid bean agar medium at 180 rpm, 28oC for 5 days. Soybean seeds were sterilized with 1% sodium hypochlorite (NaOCl) solution for 5 minutes, and then the seeds were washed with sterile water until the odor disappeared [36]. Before sowing, the seeds were inoculated with a suspension of bacteria with a cell titer of 1×108 cfu/ml for two hours at a temperature of 23oC and were sown in 500 ml vegetative vessels (three seeds per vessel). As substrate for the growth of plants, vermiculite was used, and for feeding the seedlings, Knop's solution was used (g/l) (Ca(NO3)2×4H2O: 1.00, MgSO4×7H2O: 0.25, KH2PO4: 0.25, KCl: 0.12, FeCl3×6 H2O: 0.004 g, sterile water: 1,000 ml) [37]. Before setting up the experiment, the substrate and Knop's solution were sterilized, and sterile tap water was used for watering the plants. The control consisted of soybean seeds without rhizobia inoculation. All experiments were performed in five repetitions.
2.4. Nitrogenase Activity of Rhizobia
The nitrogenase activity of rhizobia was determined by the acetylene reduction assay (ARA) [38]. For this purpose, the studied bacteria were grown on Ashby's medium under aerobic conditions to a concentration of 108 cfu/ml. Acetylene was injected into vessels with cultures to a concentration of 10% (by volume). After incubation of cultures for 1.5 hours in an acetylene atmosphere, gas samples were taken with a 1 ml syringe from a vessel, and the presence of ethylene was determined on an Agilent Technology 7890 B gas chromatograph (USA) with a flame ionization detector [39]. The nitrogenase activity was calculated using the following formula:
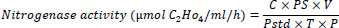 |
Where:
C = Concentration of ethylene in µmol;
PS = Peak height of the sample;
V = Volume of air space in the assay vial;
Pstd = Peak height of the standard;
T = Time of incubation in h;
P = Protein concentration of bacterial cell in mL.
2.5. Isolation of Phosphate-Solubilizing Bacteria
Phosphate-solubilizing bacteria were isolated from the rhizosphere of healthy soybean plants of the Eureka variety from the Almaty region of Kazakhstan. Rhizosphere soil samples were collected in sterile bags in compliance with the rules of asepsis. The isolation of phosphate-solubilizing bacteria was carried out as follows: 10 g of a soil sample was crushed and placed in 10 ml of sterile saline solution (0.9% NaCl) [40]. Then, we prepared dilutions from 10-1 to 10-6 and sowed them in Petri dishes with an NBRIP (National Botanical Research Institute's phosphate growth medium) agar medium with the following composition, g/l: glucose: 10.0, Ca3(PO4)2: 5.0, MgCl2×6H2O: 5.0, MgSO4×7H2O: 0.25, KCl: 0.2, (NH4)2SO4: 0.1 [41]. Each dilution was sown in three repetitions. The dishes were incubated at 28°C until transparent zones (halo zones) appeared in the environment around bacterial colonies (2-3 days). Pure bacterial cultures were obtained by picking individual colonies with the help of an inoculation loop [40]. The bacteria were cultured on a liquid and NBRIP agar medium on a shaker at 180 rpm and a temperature of 28°C.
2.6. Phosphate-Solubilizing Activity
The study of bacteria concerning the solubilization of phosphates was carried out according to the modified Segi's method [42]. The strains of phosphate-solubilizing bacteria were grown on an NBRIP medium containing insoluble tricalcium phosphate in the form of fine sediment, which gives uniform turbidity to the nutrient medium. The NBRIP agar medium was placed into the Petri dishes. After solidifying the NBRIP medium, holes with a diameter of 7 mm were punched with a sterile borer, and 0.5 ml bacterial suspension at 107 cells/ml concentration was introduced into the holes. As a negative control, sterile tap water was poured into holes. The Petri dishes were incubated at 28°C for 2-3 days until the formation of transparent zones (halo) around the holes. The phosphate-solubilizing activity of bacteria was quantified by the diameter of thalo zones in mm. All experiments were carried out in 5 replicates.
2.7. PGP Activity of Bacteria
To study the effect of bacteria on seed germination and stimulation of soybean growth, rhizobia were grown on the bean agar medium at 28°C, 180 rpm for 5 days, and phosphate-solubilizing bacteria on the NBRIP medium at 28°C, 180 rpm for 3 days. The seeds were inoculated with a suspension of bacteria with a titer of 108 cfu/ml before sowing for two hours at 23°C, at the rate of 5 ml per seed. The treated seeds were sown in 500 ml vegetative vessels. Vermiculite was used as a substrate for plant growth. Knop's solution was used to feed the seedlings. The substrate and Knop's solution were sterilized. Sterile tap water was used to water the plants. The negative control was seeds treated with sterile water [40].
Seed germination was determined by counting the number of germinated seeds from treated and control pots. To assess the growth-promoting effect of bacteria, the stem and root length were measured, and the number of leaves per plant was counted and compared with the control. The experiments were carried out in 5 replications of 3 plants per vessel. The experiment was repeated six times. A total of 150 soybean plants were used. After 30 days, morphometric traits were measured [43].
Plants were grown in a climate chamber (Memmert HPP 750 Constant Climate Chamber, Germany) under the following conditions: daylight: 12 hours, temperature: 25°C, illumination: cold white light at 6,500 K, warm light at 2,700 K; night mode: 12 hours; temperature 15°C, humidity: 65% [40].
The experiments on the influence of consortia on the development of soybean plants grown in the Almaty region were carried out in vegetative vessels (5,000 ml). The soil type was ordinary gray soil. Agrochemical indicators were humus 1.2%, easily hydrolyzed nitrogen: 58.2 mg/kg of soil, mobile phosphorus: 21.4 mg/kg of soil, and mobile potassium: 460 mg/kg. The reaction nature of the soil was slightly alkaline (pH 7.8) [40]. In the experiments, we used consortiums that included rhizobia strains (Bradyrhizobium lupini RH-6; Br. lupini RH-7) and strains of phosphate-solubilizing bacteria (Pseudomonas koreensis FT-4; Enterobacter hormaechei subsp. xiangfangensis FY-3) in various combinations. The bacteria were grown on liquid nutrient media: rhizobia on the bean agar medium at 28°C, 180 rpm for 5 days, and phosphate-solubilizing bacteria on the NBRIP medium at 28°C, 180 rpm for 3 days. The seeds of soybean varieties Eurika, Zhansaya, and Perizat were inoculated with a suspension of bacteria with a titer of 108 cfu/ml before sowing for two hours at 23°C at the rate of 5 ml per seed. The negative control was seeds treated the sterile water. The experiments were carried out in a 5-fold replication of 3 plants per vessel for 60 days. The experiment was repeated six times. A total of 150 soybean plants were used. Morphometric features were measured in two stages of soybean growth: the three unfolded trifoliate leaves stage and the full bloom stage. The experiments were carried out in open sites under field conditions.
2.8. Identification of Bacteria
The bacteria were identified by the Sanger sequencing of the 16S rRNA gene. Genomic DNA was isolated from a 24-hour-old culture of bacteria using the PureLink® Genomic DNA Kits (Invitrogen, USA). The bacteria were identified by studying the sequence of the 16S rRNA gene region with species-specific primers [40, 44, 45].
The amplification reaction mixture consisted of 12.5 μl of Q5® Hot Start High-Fidelity 2X Master Mix, 1.25 μl of each primer (10 μM), 0.03 µg of DNA, and 8.5 μl of sterile water for molecular studies [40]. The total volume of the PCR mixture was 25 μl. PCR amplification was performed in an Eppendorf ProS thermal cycler (Eppendorf, Hamburg, Germany 2012) with the following amplification mode: the temperature of denaturation: 98°C, 30 sec; annealing: 55°C, 1 min; elongation: 72°C, 40 sec, only 30 cycles; termination of the reaction: 72°C, 10 min. The amplification results were analyzed on a 1.2% agarose gel and purified using the CleanSweep ™ PCR purification reagent (Applied Biosystems, USA) [40].
Sequencing of 16S rRNA gene fragments of bacteria was performed on an automatic sequencer 3500 DNA Analyzer (Applied Biosystems, USA) using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) according to the manufacturer's protocol (BigDye® Terminator v3.1 Cycle Sequencing Kit Protocol, Applied Biosystems, USA). The sequencing results were processed using the SeqA software (Applied Biosystems). The search for homologous nucleotide sequences of 16S rRNA genes was carried out using the BLAST program (Basic Local Alignment Search Tool) in the GenBank of the International National Center for Biotechnological Information (NCBI) Database, USA [40, 46]. Phylogenetic analysis was performed using the MEGA6 software. Alignment of nucleotide sequences was performed using the ClustalW algorithm. To construct phylogenetic trees, the NJ (Neighbor-Joining) method was used.
2.9. Biocompatibility of Bacteria
To study the biocompatibility of rhizobia and phosphate-solubilizing bacteria, the perpendicular streak technique was used [47]. The studied rhizobia were sown on the surface of the bean agar medium in a Petri dish with a streak in the center. The dishes were incubated in a thermostat at 28°C. After the growth was completed, cultures of phosphate-solubilizing bacteria (test cultures) were sown with streaks perpendicular to the grown streak, starting from the edge of the dish. The dishes were incubated in an incubator at 28°C for 48 hours. In the case of the two study micro-organisms being antagonistic, their streaks appeared at a distance from each other, showing a clear zone of inhibition. Microorganisms that did not have antagonism grew near the central streak.
3. RESULTS
To isolate rhizobia, a large number of nodules from the roots of 67 healthy and large plants of the Eureka variety with well-developed root systems (large root diameters, long specific root length, and considerable root length density) were collected. Strains were isolated from the nodules, they were purified, and 24 pure rhizobia cultures were obtained. When growing on the bean agar medium, rhizobia formed colorless or milky-white mucous colonies, while on the slant agar, they formed transparent mucous, flowing down streak colonies.
To select the most active rhizobia, their nodulating and nitrogen-fixing activity was studied. The soybean variety Eureka was used in the experiments. Table 1 shows the data of the seven strains with the highest results.
| Strains of Rhizobium |
Number of Nodules Per Plant (pcs) |
The Dry Weight of Nodules (mg/plant) |
Root Dry Weight* (g/plant) |
Nitrogenase Activity (µmol C2H4/ml/h) |
|---|---|---|---|---|
| Control | 0 | 0 | 7.3 ± 0.1 | 0.00 ± 0.00 |
| MA-1 | 12.8 ± 1.2* | 128.3 ± 0.05* | 11.3 ± 0.2 | 3.87 ± 0.01** |
| RH-2 | 14.6 ± 1.4* | 130.2 ± 0.03* | 10.9 ± 0.1 | 2.32 ± 0.01** |
| RH-3 | 12.1 ± 1.1* | 142.2 ± 0.08* | 9.7 ± 0.2 | 5.25 ± 0.01** |
| RH-4 | 17.8 ± 1.4* | 150.5 ± 0.07* | 12.2 ± 0.1 | 6.45 ± 0.02** |
| RH-6 | 18.2 ± 1.3* | 153.2 ± 0.1* | 12.6 ± 0.1 | 6.71 ± 0.01** |
| RH-7 | 18.4 ± 1.1* | 158.5 ± 0.08* | 12.8 ± 0.2 | 6.74 ± 0.02** |
| RH-8 | 18.1 ± 1.4* | 157.4 ± 0.1* | 12.7 ± 0.2 | 6.37 ± 0.01** |
Rhizobia strains actively formed nodules on soybean roots and fixed atmospheric nitrogen (Table 1). It was noted that in all the inoculated plants, the nodules were pink, indicating active fixation of atmospheric nitrogen since the pink color of the nodules indicates the presence of leghemoglobin, which controls the flow of oxygen to bacteria. In the samples inoculated with MA-1, RH-2, and RH-3 strains, the number of nodules per plant ranged between 12-14 pieces, and this number increased to 17-18 in plants inoculated with RH-4, RH-6, RH-7, and RH-8 strains. Particularly, the nodules formed by these last strains were larger and darker, appeared earlier, and with higher dry weight and higher nitrogen fixation activity than those from the other strains (Table 1).
To isolate phosphate-solubilizing bacteria, 56 soil samples were collected from the rhizosphere of healthy soybean plants, and 32 pure cultures of bacteria were isolated from them. The initial screening of bacteria allowed us to select 12 strains that formed a large clearing zone in a shorter period (Fig. 1)
| Strains of Bacteria | Duration of Cultivation | |||||
|---|---|---|---|---|---|---|
| 3 Days | 5 Days | |||||
| Titer (CFU/ml) |
pH | Diameter of the Halo Zone (mm) | Titer (CFU/ml) |
pH | Diameter of the Halo Zone (mm) | |
| FL-2 | 3.3×106 | 5.1±0,1 | 18.7±1.0 | 2.7×108 | 4.5±0.06 | 25.9±1.7 |
| FY-3 | 3.5×107 | 4.9±0.1 | 22.7±1.2 | 2.8×108 | 4.4±0.04 | 26.4±1.7 |
| FT-1 | 4.3×107 | 4.6±0.08 | 19.5±1.1 | 2.5×108 | 4.4±0.05 | 26.1±1.9 |
| FT-4 | 4.4×107 | 4.5±0.05 | 23.8±1.3 | 2.6×108 | 4.3±0.06 | 26.3±1.3 |
| FM-2 | 3.3×106 | 4.7±0.07 | 17.9±1.0 | 2.8×107 | 4.5±0.04 | 18.7±1.0 |
| FM-9 | 4.0×107 | 4.5±0.04 | 16.7±0.08 | 2.4×107 | 4.5±0.03 | 19.4±1.1 |
| FC-11 | 4.1×106 | 4.6±0.08 | 16.9±0.09 | 3.4×107 | 4.4±0.04 | 19.8±1.2 |
When selecting phosphate-solubilizing bacteria, one of the important criteria, in addition to the degree of solubilization, is the rate of solubilization of phosphates into soluble forms available to plants. In this regard, the study of the phosphate-solubilizing activity of 12 selected strains was carried out in dynamics. Table 2 shows the data of the seven bacterial strains with the highest results.
Table 2 shows that the bacteria had a high ability to solubilize phosphates. It was found that four bacterial strains, FL-2, FY-3, FT-1, and FT-4, had the highest activity. The diameter of the phosphate dissolution zones by these strains fluctuated within 25.9 ± 1.7-26.4 ± 1.7 mm (fifth day of cultivation). Significant differences in the change in pH of the cultivation medium in these strains were not revealed. However, it was found that the cell titer of the strains was an order of magnitude higher than that of the others, which may be due to the higher rate of biomass accumulation by the strains. Strain FY-3 (26.4 ± 1.7 mm) showed the maximum activity, followed by FT-4 (26.3 ± 1.3 mm).
The results of the PGP activity of the selected rhizobia and phosphate-solubilizing bacteria are shown in Fig. (2).
However, according to the results of the PGP activity, the bacterial strains differed from the control. We showed that when seeds were inoculated with phosphate-solubilizing bacteria, the length of the stems increased compared to the control (stem 10.2 ± 0.5 cm; root 7.1 ± 0.2 cm) and fluctuated within 14.8 ± 0.9-20.4 ± 1.1 cm, that of the roots –9.9 ± 0.2-10.8 ± 0.4 cm, whereas when inoculated with rhizobia, the length of the stems increased – 16.4 ± 1.0-23.7 ± 1.3 cm, that of the roots – 10.9 ± 1.0-12.8 ± 1.4 cm. From the given data, it shows that the growth-stimulating activity of rhizobia was slightly higher than that of phosphate-solubilizing bacteria. Rhizobia also increased the germination of soybean seeds up to 98.4 ± 1.7% and phosphate-solubilizing bacteria up to 83.3 ± 1.5%; in control, the germination of seeds was 58.6 ± 1.3%. Data are average values of five replications ± SEM at p < 0.05.
The biocompatibility of rhizobia and phosphate-solubilizing bacteria was studied using dual cultures on a solid nutrient medium. A strain of rhizobia was seeded on the surface of the agar medium with a vertical stroke in the center. After completion of growth, phosphate-solubilizing bacteria were inoculated with horizontal strokes perpendicular to the grown streak. Next, an analysis of their growth was carried out. In antagonistic interaction, the growth of phosphate-solubilizing bacteria was at some distance from the vertical streak of rhizobia (Fig. 3).

Vertical stroke in the center – rhizobia strain RH-7 (a) and RH-4 (b), horizontal strokes – phosphate-solubilizing bacteria.
The biocompatibility study made it possible to select rhizobia and phosphate-solubilizing bacteria with no antagonism (Fig. 3).
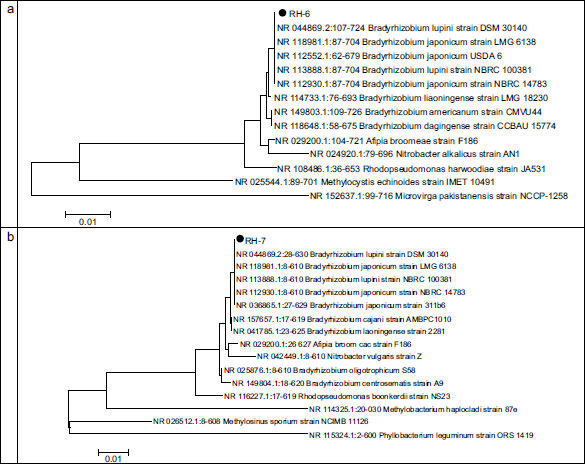
According to these results, two rhizobia strains (RH-6, RH-7) and two phosphate-solubilizing bacterial strains (FY-3, FT-4) were selected to set consortia. These strains were previously characterized by sequencing (603 bp) the 16 rDNA region. Figs. (4 and 5) show the relative phylograms.
Fig. (4) shows that, phylogenetically, the FT-4 strain is closest to the strains of the genus Pseudomonas. The maximum similarity was reported with the nucleotide sequence of the 16S gene of strain NR 25228.1:65-770 Pseudomonas koreensis strain Ps 9-14 from the GenBank database (percentage of identity was 99.72%). The FY-3 strain is on the same branch as the reference strains of the GenBank database of the genus Enterobacter. Percentage of identity with the reference strain NR 126208.1:15-727 Enterobacter hormaechei subsp. xiangfangensis strain 10-17 equals 99.58%.
Fig. (5) shows the dendrograms of rhizobia, RH-6, and RH-7 strains.
Fig. (5) shows that phylogenetically strains RH-6 and RH-7 are closest to Bradyrhizobium and are on the same branch as the reference strain from the GenBank NCBI database NR 044869.2 Bradyrhizobium lupini strain DSM 30140. The identity percentage with this strain equals 99.98%.
Nucleotide sequences of bacterial strains are given in the supplementary material of the article.
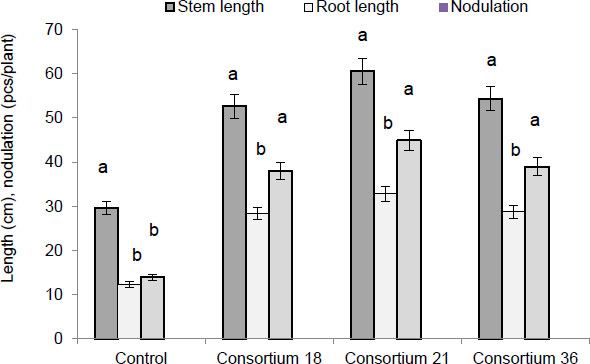
Based on selected strains of rhizobia and phosphate-solubilizing bacteria, consortia of agronomically important microorganisms were created, and their effect on the Eureka variety soybean plants’ growth and root nodulation was studied. Fig. (6) shows data on the impact of the three most effective consortia: (No 18 (strains RH-6 and FY-3), No 21 (strains RH-7 and FT-4), and No 36 (strains RH-7 and FY-3).
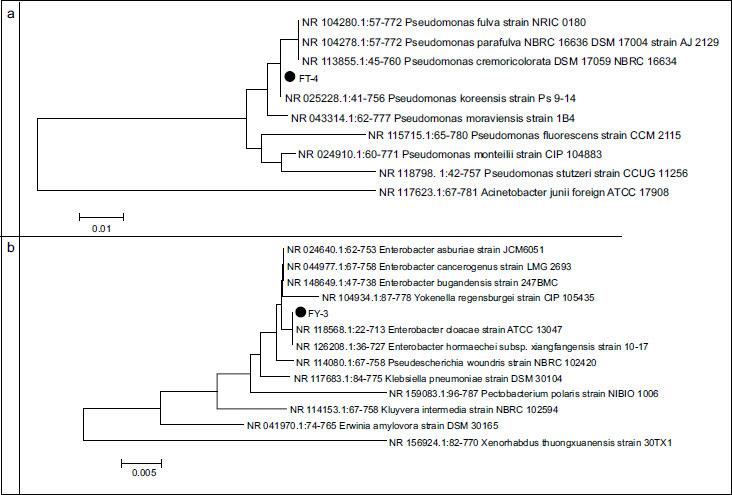
Fig. (6) shows that the consortia had a high PGP activity than the control. In seed inoculation by consortia, the length of the soybean stems increased compared to the control (stem 29.7 ± 1.8 cm; root 12.3 ± 1.2 cm; nodule 14.2 ± 1.3 cm) and fluctuated within 52.6 ± 0.9-60.5 ± 3.1 cm, the roots – 28.4 ± 0.2-32.8 ± 2.4 cm, and the number of nodules per plant – 38.2 ± 1.5-45.1 ± 2.3 cm. Data are average values of five replications ± SEM at p < 0.05.
When soybean seeds of the Zhansaya and Perizat varieties were inoculated with the consortiums, we obtained results that correlated with the results of the Eureka soybean variety. The differences were in numerical terms; therefore, data for these varieties were not provided. Results demonstrated that the consortia used in our study were effective. It was found that Consortium No. 21 was the most effective for all three soybean varieties. During the pre-sowing treatment of seeds of the Eureka variety, the stem length was 60.5 ± 3.1 cm, the root length was 32.8 ± 2.4 cm, and the number of nodules per plant was 45.1 ± 2.3 cm (control: stem 29.7 ± 1.8 cm; root 12.3 ± 1.2 cm; nodule 14.2 ± 1.3 cm). These indicators for the Zhansaya variety were: stem length was 58.6 ± 2.4 cm, root length was 30.8 ± 2.1 cm, and the number of nodules per plant was 42.6 ± 1.3 cm (control: stem 25.7 ± 1.5 cm; root 11.7 ± 1.1 cm; nodule 12.2 ± 0.9 cm); for the Perizat variety, stem length was 57.8 ± 2.0 cm, root length was 28.3 ± 1.8 cm, and the number of nodules per plant was 41.1 ± 1.2 cm (control: stem 24.6 ± 1.1 cm; root 10.8 ± 0.9 cm; nodule 11.7 ± 1.0 cm). Data are average values of five replications ± SEM at p < 0.05.
The effect of consortia on nitrogen fixation, phosphate solubilization, and protein accumulation in the groundmass of soybeans has been studied. Since it was planned to use the created consortiums in production conditions in the fields, the soil taken from the field of the Almaty region of Kazakhstan was used in the experiments, which created conditions closer to field conditions.
Table 3 shows that inoculation of soybean seeds by consortia significantly increases nitrogen fixation by soybean nodules, phosphate solubilization, and the protein content in the dry mass of plants (by 1.4-1.9 times) compared to the control. The formation of nodules on the roots of soybean plants in control was explained by the fact that the soil from fields of the Almaty region of Kazakhstan, which contained rhizobia, was used for the experiments. Consortium No. 21 was the most effective one. The consortium included rhizobia Bradyrhizobium lupini RH-7 and phosphate-solubilizing bacteria Pseudomonas koreensis FT-4.
| Variants | Nitrogenase Activity** (µmole C2H4/g Nodules /h) | Protein Ground Mass/Plant* (%) |
Halo Zone Diameter (mm) |
|---|---|---|---|
| Eureka Variety | |||
| Control | 11.75 ± 0.7 | 11.4 ± 0.8 | 0 |
| Consortium No 18 | 28.34 ± 1.1 | 18.7 ± 1.6 | 32.5 ± 2.2* |
| Consortium No 21 | 34.84 ± 1.3 | 21.5 ± 2.0 | 35.6 ± 2.1* |
| Consortium No 35 | 29.76 ± 1.3 | 19.8 ± 1.5 | 31.2 ± 2.0* |
| Zhansaya Variety | |||
| Control | 10.22 ± 0.9 | 11.6 ± 0.6 | 0 |
| Consortium No 18 | 24.53 ± 1.4 | 16.3 ± 1.2 | 31.9 ± 1.8* |
| Consortium No 21 | 32.64 ± 1.3 | 19.8 ± 1.5 | 33.5 ± 2.0* |
| Consortium No 35 | 27.16 ± 1.2 | 17.4 ± 1.2 | 28.2 ± 1.9* |
| Perizat Variety | |||
| Control | 10.47 ± 0.8 | 10.9 ± 0.7 | 0 |
| Consortium No 18 | 24.18 ± 1.0 | 15.9 ± 1.0 | 29.8 ± 1.7* |
| Consortium No 21 | 29.34 ± 1.3 | 20.3 ± 1.2 | 34.8 ± 2.2* |
| Consortium No 35 | 26.14 ± 1.5 | 18.5 ± 1.4 | 30.1 ± 1.7* |
4. DISCUSSION
To obtain the high productivity of crops and especially legumes, basic nutrients, such as nitrogen, phosphorus, and potassium, are needed. When growing soybeans, mineral nitrogen and phosphorus fertilizers are used. The use of high doses of fertilizers makes it possible to achieve large yields but leads to negative consequences, including a decrease in fertility, a decrease in the biodiversity of agrocenoses, and the accumulation of chemicals in food, which affects people's health. Organic fertilizers are an alternative to chemical fertilizers that can mitigate these problems. They gradually release nutrients, maintain the balance of basic elements in the soil, are effective sources of nutrition for soil microbes, and improve their structure. Therefore, the provision of nitrogen and phosphorus by plants without the use of mineral fertilizers is one of the main problems of organic agriculture [50]. This problem can be solved through the use of biofertilizers based on agronomically important microorganisms. Inoculation of soybeans with effective strains of rhizobia and phosphate-solubilizing bacteria not only reduces the use of mineral fertilizers but also makes nitrogen and phosphorus fertilization unnecessary [51, 52]. However, there are several problems when using biofertilizers. One of them is the use of exogenous strains which might not compete with the indigenous populations of rhizosphere microbes; the other is the incompatibility of strains that are part of an association or consortium.
Earlier, it was found that the joint use of rhizobia and rhizospheric bacteria positively affected the growth of different crops [53-55]. However, the combined effect of rhizobia and phosphate-solubilizing bacteria on soybean growth has not been sufficiently studied [56]. Thus, it is of interest to create and evaluate the effectiveness of a consortium of agronomically important microorganisms for growing soybean, including rhizobia and phosphate-mobilizing bacteria. Screening of local bacteria already adapted to local conditions and searching for highly effective strains for use as inoculants are promising strategies to increase soybean productivity.
In our study, strains of rhizobia (24 strains) and phosphate-solubilizing bacteria (32 strains) adapted to the soil and climatic conditions of the Almaty region in Kazakhstan were isolated from the nodules and rhizosphere of soybean. Their biological activity was studied, and highly active bacterial strains were selected. It was found that all the selected strains had PGP activity and stimulated the growth of soybean plants; rhizobia actively fixed air nitrogen and formed nodules on soybean roots, and phosphate-solubilizing bacteria actively absorbed phosphates. Identification of bacteria by sequencing the 16S rRNA gene showed that phosphate-solubilizing bacteria belonged to the genera Pseudomonas and Enterobacter and rhizobia to the genus Rhizobium. These bacteria have been successfully used as soybean bio-inoculants to increase its yield [57-59].
We studied the biocompatibility of rhizobia and phosphate-mobilizing bacteria and selected strains without antagonistic activity toward each other. Based on these strains, consortia were created. Our study showed that the use of consortia increased the growth of soybean stems and roots, the number of nodules on soybean roots, and their nitrogen-fixing ability, as well as the protein content in soybean plants. The positive effect of consortium inoculation on these plant traits was noted in all soybean varieties investigated. The best-performing consortium was made up of rhizobia Bradyrhizobium lupini RH-7 and phosphate-solubilizing bacteria Pseudomonas koreensis FT-4.
CONCLUSION
The results of our study demonstrated the high prospects of using a consortium of rhizobia and phosphate-solubilizing bacteria for growing soybean. It was found that under the influence of consortium, the nitrogen fixation of plant nodules, the protein content in soybean plants, and the absorption of phosphorus increased. These likely result from the nitrogen provided by rhizobia and phosphorus solubilization by phosphate-solubilizing bacteria. The consortium was based on local strains. We believe that inoculation with indigenous bacteria, which are adapted to specific local conditions, is a possible strategy for increasing soybean productivity on a local scale. Hence, it is important to offer new approaches to increase crop productivity sustainably.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This work was carried out with the financial support of the Ministry of Education and Science of Kazakhstan within the framework of the grant project IRN АP09259080.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.
SUPPLEMENTAL MATERIAL
Additional data on the results of strain sequencing:
Nucleotide sequence of strain FY-3 – Enterobacter hormaechei subsp. xiangfangensis FY-3:
TGACGAGTGGCGGACGGGTGAGTAATGTCTGGGAAACTGCCTGATGGAGGGGGATAACTACTGGAAACGGTAGCTAATACCGCATAACGTCGCAAGACCAAAGAGGGGGACCTTCGGGCCTCTTGCCATCGGATGTGCCCAGATGGGATTAGCTAGTAGGTGGGGTAACGGCTCACCTAGGCGACGATCCCTAGCTGGTCTGAGAGGATGACCAGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTATGAAGAAGGCCTTCGGGTTGTAAAGTACTTTCAGCGGGGAGGAAGGCGACAGGGTTAATAACCCTGTCGATTGACGTTACCCGCAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAATACGGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAACGCACGCAGGCGGTCTGTCAAGTCGGATGTGAAATCCCCGGGCTCAACCTGGGAACTGCATTCGAAACTGGCAGGCTAGAGTCTTGTAGAGGGGGGTAGAATTCCAGGTGTAGCGGTGAAATGCGTAGAGATCTGGAGGAATACCGGTGGCGAAGGCGGCCCCCTGGACAAAGACTGACGCTCAGGTGCGAAAGCGTGGGGAGCAAACAGGA
Nucleotide sequence of strain FT-4 – Pseudomonas koreensis FT-4:
GAGAGGAGCTTGCTCCTGGATTCAGCGGCGGACGGGTGAGTAATGCCTAGGAATCTGCCTGGTAGTGGGGGACAACGTTTCGAAAGGAACGCTAATACCGCATACGTCCTACGGGAGAAAGCAGGGGACCTTCGGGCCTTGCGCTATCAGATGAGCCTAGGTCGGATTAGCTAGTTGGTGAGGTAATGGCTCACCAAGGCGACGATCCGTAACTGGTCTGAGAGGATGATCAGTCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGGGAATATTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTTCGGATTGTAAAGCACTTTAAGTTGGGAGGAAGGGTTGTAGATTAATACTCTGCAATTTTGACGTTACCGACAGAATAAGCACCGGCTAACTCTGTGCCAGCAGCCGCGGTAATACAGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTAGGTGGTTTGTTAAGTTGGATGTGAAATCCCCGGGCTCAACCTGGGAACTGCATCCAAAACTGGCAAGCTAGAGTATGGTAGAGGGTGGTGGAATTTCCTGTGTAGCGGTGAAATGCGTAGATATAGGAAGGAACACCAGTGGCGAAGGCGACCACCTGGACTGATACTGACACTGAGGTGCGAAAGCGTGGGGAGCAAACAGGATTAGAT
Nucleotide sequence of strain RH-6 – Bradyrhizobium lupini RH-6:
ACGTACCTTTTGGTTCGGAACAACACAGGGAAACTTGTGCTAATACCGGATAAGCCCTTACGGGGAAAGATTTATCGCCGAAAGATCGGCCCGCGTCTGATTAGCTAGTTGGTGAGGTAATGGCTCACCAAGGCGACGATCAGTAGCTGGTCTGAGAGGATGATCAGCCACATTGGGACTGAGACACGGCCCAAACTCCTACGGGAGGCAGCAGTGGGGAATATTGGACAATGGGGGCAACCCTGATCCAGCCATGCCGCGTGAGTGATGAAGGCCCTAGGGTTGTAAAGCTCTTTTGTGCGGGAAGATAATGACGGTACCGCAAGAATAAGCCCCGGCTAACTTCGTGCCAGCAGCCGCGGTAATACGAAGGGGGCTAGCGTTGCTCGGAATCACTGGGCGTAAAGGGTGCGTAGGCGGGTCTTTAAGTCAGGGGTGAAATCCTGGAGCTCAACTCCAGAACTGCCTTTGATACTGAGGATCTTGAGTTCGGGAGAGGTGAGTGGAACTGCGAGTGTAGAGGTGAAATTCGTAGATATTCGCAAGAACACCAGTGGCGAAGGCGGCTCACTGGCCCGATACTGACGCTGAGGCACGAAAGCGTGGGGAGCAAACAGG
Nucleotide sequence of strain RH-7 – Bradyrhizobium lupini RH-7:
GCTGGCGGCAGGCTTAACACATGCAAGTCGAGCGGGCGTAGCAATACGTCAGCGGCAGACGGGTGAGTAACGCGTGGGAACGTACCTTTTGGTTCGGAACAACACAGGGAAACTTGTGCTAATACCGGATAAGCCCTTACGGGGAAAGATTTATCGCCGAAAGATCGGCCCGCGTCTGATTAGCTAGTTGGTGAGGTAATGGCTCACCAAGGCGACGATCAGTAGCTGGTCTGAGAGGATGATCAGCCACATTGGGACTGAGACACGGCCCAAACTCCTACGGGAGGCAGCAGTGGGGAATATTGGACAATGGGGGCAACCCTGATCCAGCCATGCCGCGTGAGTGATGAAGGCCCTAGGGTTGTAAAGCTCTTTTGTGCGGGAAGATAATGACGGTACCGCAAGAATAAGCCCCGGCTAACTTCGTGCCAGCAGCCGCGGTAATACGAAGGGGGCTAGCGTTGCTCGGAATCACTGGGCGTAAAGGGTGCGTAGGCGGGTCTTTAAGTCAGGGGTGAAATCCTGGAGCTCAACTCCAGAACTGCCTTTGATACTGAGGATCTTGAGTTCGGGAGAGGTGAGTGGAACTGCGAGTGTAGAGGT


