All published articles of this journal are available on ScienceDirect.
Screening of the Bread Wheat Varieties for the Leaf Rust Resistance Gene Lr34/Yr18/Sr57/Pm38/Bdv1
Abstract
Background:
The allelic composition of the gene Lr34/Yr18/Sr57/Pm38/Bdv1, which is associated with resistance to leaf rust in varieties of bread wheat (Triticum aestivum L.), has been investigated.
Methods:
Three DNA markers were used to determine the allelic state of the gene Lr34/Yr18/Sr57/Pm38/Bdv1: the co-dominant molecular genetic markers cssfr5 and csLV34 and the microsatellite marker Xgwm295.
Results:
Among 32 cultivars evaluated for resistance to leaf rust, 4 were highly resistant, 26 were resistant and 2 were moderately susceptible. Using the co-dominant marker cssfr5 based on the detection of the polymorphic state of one of the exons of the gene Lr34/Yr18/Sr57/Pm38/Bdv1, the Lr34(+) allele, which confers resistance to leaf rust, was found in 25% of the studied varieties. The coincidence between the results obtained with the markers cssfr5 and csLV34 was 84.5%.
Conclusion:
The data of the conducted molecular genetic analysis were supplemented by observations of the resistance of the studied varieties to leaf rust in the field. The obtained data can be used in breeding programs to develop new varieties and breeding lines with leaf rust resistance.
1. INTRODUCTION
Leaf rust caused by the pathogenic fungus Puccinia triticina Erikss. is one of the most widespread wheat diseases in Ukraine, as well as globally [1, 2]. According to FAO, during rust epiphytotic, wheat crop losses can amount up to 40%. The most effective method of plant protection is to develop resistant varieties. At present, more than 80 genes for leaf rust resistance have been identified in the genome of wheat, and its relatives [3], and molecular markers are commonly used to identify them [4]. Leaf rust resistance genes were designated as Lr in the catalogue of gene symbols [3].
Wheat breeding for resistance to fungal diseases is based on a combination of genes of race-specific (vertical) and race-nonspecific (horizontal) resistance, which makes it possible to ensure a high level of resistance over a long period of time [5]. Most genes that control race-specific resistance to rusts remain effective only for several years [6]. Race-nonspecific rust resistance is commonly moderate, but this type of resistance, which is not based on specific recognition between the host and the pathogen, proves to be durable. Horizontal resistance to rust manifests at the adult plant stage as the adult plant resistance (APR) [7]. Unlike most Lr genes, which provide resistance for only a few years, the Lr34/Yr18/Sr57/Pm38/Bdv1 gene retains its effectiveness for many seasons [8]. The Lr34/Yr18/Sr57/Pm38/Bdv1 gene is race-nonspecific and provides moderate resistance to leaf rust in adult plants [1, 9]. This gene was first discovered in 1977 and mapped to chromosome 7D [10]. The Lr34 gene also provides moderate resistance to other biotrophic pathogens: yellow rust caused by P. striiformis Westend. f. sp. tritici (in this case, it is referred to as the Yr18 gene) [11], stem rust caused by Puccinia graminis Pers. (as Sr57) and powdery mildew caused by Blumeria graminis (DC.) (DC.) Speer f. sp. tritici (referred to as Pm38) [12]. The Lr34/Yr18/Sr57/Pm38 pleiotropic set possessed by Bezostaya 1 currently represents an important target for wheat breeding because it is now amenable to molecular selection [2]. It is also associated with resistance to yellow dwarf barley mosaic virus (the Bdv1 gene) [13]. Besides, the Lr34/Yr18/Sr57/Pm38/Bdv1 gene is also closely linked to the Ltn1 gene for leaf tip necrosis [14]. The phenotypic expression of the Ltn1 gene was used by Singh et al. [15] to detect the Lr34/Yr18/Sr57/Pm38/Bdv1 gene, but the multigenic effect on the overall manifestation of flag leaf tip necrosis and unstable Ltn1 expression under different conditions may lead to ambiguous results [16].
There is considerable interest in developing effective methods for detecting wheat disease resistance genes. Microsatellite DNA markers Xgwm295 and Xgwm1220 were among the first markers used to identify the Lr34/Yr18/Sr57/Pm38/Bdv1 gene [17]. However, these markers have not been widely used because of their low efficiency. In addition, two other markers, Xswm10 and csLVMS1 [18], were used to detect the Lr34/Yr18/Sr57/Pm38/Bdv1 gene. However, practical application of these markers was restrained by a difficulty in routine differentiation of alleles due to the small difference between the amplicons of 206 bp and 208 bp for the marker Xswm10 [12], and similarly, between the amplicons of 224 bp and 226 bp for the marker csLVMS1 [19]. The most widely used molecular marker for Lr34/Yr18/Sr57/Pm38/Bdv1 gene is csLV34 [11]. This co-dominant marker was actively used to detect alleles of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene in bread wheat samples from Europe, Australia, Canada, and the USA [20, 21].
The nucleotide sequence of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene is 11805 bp, contains 24 exons and encodes a protein with a length of 1401 amino acid residues [22]. The alleles at this locus differ by a single nucleotide substitution in exon 4, a trinucleotide deletion in exon 11, and a single nucleotide substitution in exon 12 [8, 23]. Based on the locus structure, a co-dominant molecular genetic marker cssfr5 was developed and proved to be the most effective [8]. However, according to the literature, there are some varieties carrying a characteristic resistance allele of the marker Lr34(+) in exon 11, but they do not possess the leaf rust resistance [7].
The aim of our study was to compare the efficiency and reliability of widely used indirect molecular markers csLV34 and Xgwm295 to the locus-specific marker cssfr5 of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene in a sample of modern wheat varieties originating from different climatic zones and to evaluate the possible impact of the gene on leaf rust resistance in the field.
2. MATERIALS AND METHODS
To assess the resistance of wheat samples and identify carriers of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene, Ukrainian varieties developed at the Institute of Plant Physiology and Genetics at the National Academy of Sciences of Ukraine (IPPG NASU) under the supervision of Prof. V.V. Morgun, varieties of the Myronivka Institute of Wheat named after V.M. Remeslo at the National Academy of Agrarian Sciences of Ukraine (NAASU) (hereinafter MIW), the Plant Breeding and Genetics Institute – National Center of Seed and Cultivar Investigation (PBGI), the NAASU National Science Centre “Institute of Agriculture” (IA), the company “Roden 10”, as well as some varieties from other countries have been used.
Isolation of total DNA from plant material. Wheat grains were used as plant material for DNA isolation. The isolation of total DNA was performed by the CTAB method [24].
Polymerase chain reaction (PCR). The reaction mixture consisted of the following components: specific primers (Table 1) with the concentration of 0.5 μm, 2 μl of 10 × Reaction buffer B, 2.0 mM MgCl2, 0.2 mM each deoxyribonucleotide-3-phosphate (Thermo Fisher Scientific), 0.5 units of DreamTaq polymerase ™ DNA Polymerase (Thermo Fisher Scientific), 50-100 ng of total DNA, deionized water Milli-Q (Merck Millipore) adjusted to a final volume of 20 μl. Amplification reactions were performed in a Mastercycler gradient (Eppendorf) thermal cycler.
| S.No | Marker | Primers and Their Sequence | Amplicon Size, bp |
|---|---|---|---|
| 1. | Xgwm295 | F: GTGAAGCAGACCCACAACAC R:GACGGCTGCGACGTAGAG |
256 [25] 254 250 |
| 2. | csLV34 | csLV34F: GTTGGTTAAGACTGGTGATGG csLV34R: TGCTTGCTATTGCTGAATAGT |
229 [21] 150 [20] |
| 3. | cssfr5 | L34DINT9F: 5'TTGATGAAACCAGTTTTTTTTCTA3' L34MINUSR:5'TATGCCATTTAACATAATCATGAA3' L34SPF:5'GGGAGCATTATTTTTTTCCATCATG3' L34DINT13R2:5'ACTTTCCTGAAAATAATACAAGCA3' |
751 [8] 523 |
Leaf rust resistance of the wheat varieties was assessed at the Experimental Agricultural Production fields of IPPG NAS of Ukraine. Evaluation of the extent of the leaf rust development was performed using the integrated scale of resistance of cereals against P. triticina (Table 2).
The amplification programs used were the following (according to [8, 20, 21, 25] with modifications). For the marker cssfr5: initial denaturation at 94oC for 3 min, 8 cycles: denaturation at 94oC, annealing at 68 (-1) oC for 30 s, elongation at 72 oC for 50 s, then 26 cycles: denaturation at 94oC for 30 s, annealing at 60oC for 30 s, elongation at 72oC for 50 s, final elongation at 72oC for 5 min. For the marker csLV34: initial denaturation for 3 min at 94°C, 38 cycles: denaturation at 94°C for 45 s, annealing for 30 s at 55°C, elongation at 72°C for 1 min, final elongation at 72°C for 5 min. For the marker Xgwm295: initial denaturation at 94°C for 3 min, 35 cycles: denaturation at 94°C for 30 s, annealing at 60°C for 30 s, elongation at 72°C for 1 min, final elongation at 72°C for 5 min.
DNA electrophoresis. PCR products were separated in an agarose gel in SB buffer (5 mM Na2B4O7, pH 8.5) and LB buffer (10 mM Li2B4O7, pH 8.5). GeneRulerTM DNA Ladder Mix (Thermo Fisher Scientific), Quick-Load® Purple 50 bp DNA Ladder (BioLabs) and pUC19 / MspI (Thermo Fisher Scientific) molecular weight markers were used to determine the product size. Amplicons were visualized in ultraviolet light (LKB Transilluminator Macrovue 2011), registered with the Canon EOS 600D photosystem. The images were processed with a GIMP editor and GelAnalyzer. RunSAFE, a non-mutagenic fluorescent reagent that provides instant visualization of DNA bands under UV illumination of agarose gels, was used for electrophoresis of amplicons from PCR with the csLV34 marker.
To analyze the amplification patterns for the marker Xgwm295, we used an Agilent 2100 bioanalyzer, based on a lab-on-a-chip technology and is designed for the analysis of proteins and nucleic acids. With this bioanalyzer, the analysis time was reduced to 40 minutes. The bioanalyzer allows to separate DNA fragments with an accuracy of up to 3 base pairs.
The leaf rust resistance of bread wheat plants was assessed in the dynamics (throughout the course of the disease development). The main field assessment was carried out at the period of maximum disease development – at the milk stage [26].
The severity of the disease was determined by the area of the affected surface of leaves covered with spots or intensity of other symptoms by the formula [26]:
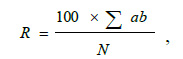
where R is the disease severity, %;
а–number of plants with disease symptoms;
b–intensity of infection, %;
N–total number of plants.
The resulting data were analyzed by one-way analysis of variance.
3. RESULTS
Initially, to identify the Lr34/Yr18/Sr57/Pm38/Bdv1 gene, we used the Xgwm295 microsatellite marker, located on chromosome 7D. Among the cultivars analyzed using the microsatellite marker Xgwm295, the following alleles were detected: 250 bp, 254 bp, 256 bp. The cultivars ‘Bogdana’, ‘Zolotokolosa’, ‘Smuglyanka’ carried the 250-bp allele. The 254-bp allele was detected in the varieties ‘Podolyanka’ and ‘Natalka’. For the rest of the varieties, the 256-bp allele was identified (Fig. 1). The previous research reported that the presence of the 254-bp allele of the microsatellite marker Xgwm295 indicates the presence of the gene Lr34/Yr18/Sr57/Pm38/Bdv1 [12].
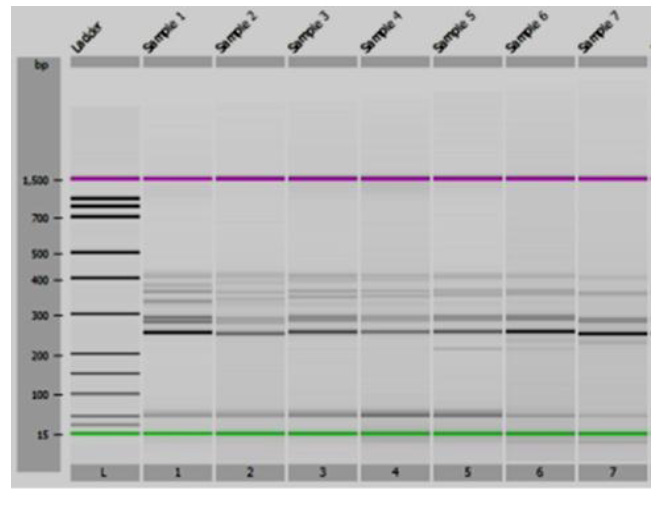
In addition, the co-dominant marker csLV34 was used. A sufficiently high degree of association of this marker with the resistance gene Lr34/Yr18/Sr57/Pm38/Bdv1 has been established [22] and verified [27, 28]. This marker detects two alleles: csLV34a with amplified fragments of 229 bp, indicating an absence of wheat resistance to leaf rust, and csLV34b – the 150-bp fragments, indicating the presence of such resistance [29]. These alleles have a deletion size of 79 bp. Only one of the two alleles of the csLV34 marker was detected in all studied wheat samples (Fig. 2). The cultivars polymorphic for this trait were not detected in the samples under study. The resistance-associated csLV34b allele (150 bp) was detected in 5 wheat varieties: ‘Zolotokolosa’, ‘Mironovskaya-30’, ‘Panna’, ‘Glenlea’, and ‘Nedra’, which accounted for 15.6% of the samples studied [30]. The csLV34a allele was present in 27 samples or 84.4% of the total number of 32 varieties tested. The full list of the cultivars analyzed is presented in Table 2.
| Score | Disease Symptoms | Degree of Resistance |
|---|---|---|
| 9 | No visible symptoms | Immune |
| 8 | Rare chlorotic and necrotic spots with very small uredinia are observed. The infection intensity 1-5% | Highly resistant |
| 7-6 | Small and moderate pustules in chlorotic and necrotic spots, the infection intensity 6-10% and 11-15% | Resistant |
| 5 | Uredinia with the infection intensity 16-25%, Light chlorosis and/or necrosis can be observed |
Moderately susceptible |
| 4-3 | Moderate to large uredinia with the infection intensity from 26-40% to 41-65%, light chlorosis is possible | Susceptible |
| 2 | Large uredinia, infection intensity 66-90% | Highly susceptible |
| 1 | Large merged uredinia, infection intensity 91-100% | Extremely susceptible |
| S.No | Cultivar | Size of the Amplified Fragments for Xgwm295 (bp) | Allelic State of the Lr34 Gene | Size of the Amplified Fragments for csLV34 (bp) | Allelic State of the Lr34 Gene | Size of the Amplified Fragments for cssfr5 (bp) | Allelic State of the Lr34 Gene | |
|---|---|---|---|---|---|---|---|---|
| Ukrainian cultivars | ||||||||
| Cultivars from the Institute of Plant Physiology and Genetics of the NAS of Ukraine | ||||||||
| 1 | ‘Bogdana’ | 250 | - | 229 | - | 523 | - | |
| 2 | ‘Vesnyanka’ | * | - | 229 | - | 523 | - | |
| 3 | ‘Volodarka’ | * | 229 | - | 751 | + | ||
| 4 | ‘Zimoyarka’ | 256 | - | 229 | - | 523 | - | |
| 5 | ‘Zolotokolosa’ | 250 | - | 150 | + | 523 | - | |
| 6 | ‘Kievskaya-Ostistaya’ | 256 | - | 229 | - | 523 | - | |
| 7 | ‘Lasunya’ | 256 | - | 229 | - | 523 | - | |
| 8 | ‘Natalka’ | 254 | + | 229 | - | 523 | - | |
| 9 | ‘Niva-Kievshciny’ | 256 | - | 229 | - | 523 | - | |
| 10 | ‘Novokiivska’ | 256 | - | 229 | - | 523 | - | |
| 11 | ‘Pereyaslavka’ | 256 | - | 229 | - | 523 | - | |
| 12 | ‘Pivna’ | 256 | - | 229 | - | 523 | - | |
| 13 | ‘Podolyanka’ | 254 | + | 229 | - | 751 | + | |
| 14 | ‘Smuglyanka’ | 250 | - | 229 | - | 523 | - | |
| 15 | ‘Sonechko’ | 256 | - | 229 | - | 523 | - | |
| 16 | ‘Favoritka’ | 256 | - | 229 | - | 523 | - | |
| 17 | ‘Khutoryanka’ | 256 | - | 229 | - | 523 | - | |
| 18 | ‘Yatran-60’ | 256 | - | 229 | - | 751 | + | |
| Cultivars from Remeslo Institute of Wheat of the NAAS of Ukraine | ||||||||
| 19 | ‘Krizhinka’ | * | 229 | - | 751 | + | ||
| 20 | ‘Mironovskaya-808’ | 256 | - | 229 | - | 523 | - | |
| 21 | ‘Mironovskaya-30’ | 256 | - | 150 | + | 751 | + | |
| Cultivars of Plant Breeding and Genetics Institute – National Center of Seed and Cultivar Investigation - of the NAAS of Ukraine | ||||||||
| 22 | ‘Bilyava’ | 256 | - | 229 | - | 523 | - | |
| 23 | ‘Oksana’ | 256 | - | 229 | - | 523 | - | |
| 24 | ‘Panna’ | 256 | - | 150 | + | 751 | + | |
| Cultivars of the National Scientific Centre “Institute of Agriculture of the NAAS of Ukraine” | ||||||||
| 25 | ‘Nedra’ | 256 | - | 150 | + | 751 | + | |
| Cultivar of the “Roden10” farm | ||||||||
| 26 | ‘Torchynska’ | 256 | - | 229 | - | 523 | - | |
| Foreign cultivars | ||||||||
| 27 | ‘Aranka’ | 256 | - | 229 | - | 523 | - | |
| 28 | ‘Glenlea’ | 256 | - | 150 | + | 751 | + | |
| 29 | ‘Granny’ | 256 | - | 229 | - | 523 | - | |
| 30 | ‘Triso’ | 256 | - | 229 | - | 523 | - | |
| 31 | ‘Tybalt’ | 256 | - | 229 | - | 523 | - | |
| 32 | ‘Federer’ | 256 | - | 229 | - | 523 | - | |
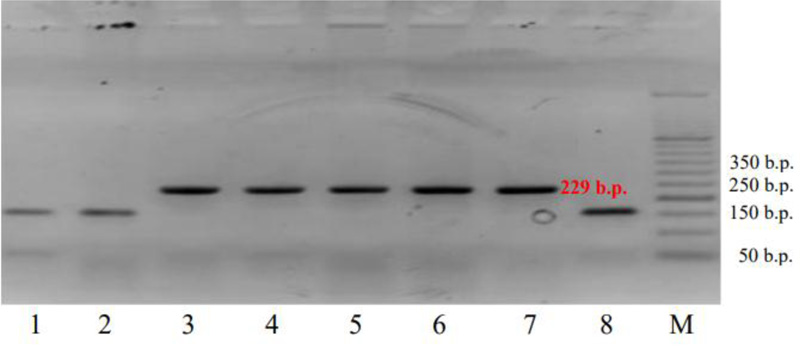
Subsequently, another study was performed to identify the allelic state of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene using the molecular genetic marker cssfr5. Fig. (3) shows an electrophoregram with the amplified DNA fragments for the wheat varieties analyzed with the marker cssfr5.
Out of the whole set of 32 varieties under study, only 8, ‘Podolyanka’, ‘Krizhinka’, ‘Yatran-60’, ‘Volodarka’, ‘Mironovskaya-30’, ‘Panna’, ‘Nedra’ and ‘Glenlea’, contained the Lr34(+) allele, which constituted 25% of the total number of the varieties tested (Table 2 and Fig. 3). 26 of them were selected in Ukraine (Table 3).
The allele Lr34(+) of the marker cssfr5 was identified only in 7 Ukrainian varieties, which is 27%, and in one out of 6 foreign varieties (‘Glenlea’). The results obtained cssfr5 were not completely identical to those with the marker csLV34 in 27 varieties, and the degree of coincidence proved to be 84.5%. Apparently, there are differences in the use of these two markers to identify the gene Lr34/Yr18/Sr57/Pm38/Bdv1.
Our data showed that in the studied wheat cultivars, resistance to leaf rust was between 5 and 8 points. The cultivars ‘Natalka’, ‘Smuglyanka’, ‘Zolotokolosa’ and ‘Favoritka’ had a resistance score of 8 and were highly resistant to leaf rust. The plants had almost no signs of the disease and the intensity of the lesion was as low as up to 5%. The other 26 varieties were less resistant, with the intensity of the lesion ranging from 5.6% to 14.8%. The most susceptible to the disease were varieties ‘Khutoryanka’ and ‘Mironovskaya-808’. The total intensity of lesion per plant for the variety ‘Khutoryanka’ was 16.7%, and for the variety ‘Mironovskaya-808’ – 24.2% (Table 4). Our data indicate (Table 4) that out of 32 studied cultivars, 4 cultivars were highly resistant, 26 – resistant and 2 – moderately susceptible.
| S.No | Cultivar | Leaf Rust | ||
|---|---|---|---|---|
| Index of Resistance | The Intensity of Lesion, % | Level of esistance or Susceptibility | ||
| Ukrainian cultivars | ||||
| Cultivars from the Institute of Plant Physiology and Genetics of NAS of Ukraine | ||||
| 1 | ‘Bogdana’ | 7-6 | 6.7 | Resistant |
| 2 | ‘Vesnyanka’ | 7-6 | 14.8 | Resistant |
| 3 | ‘Volodarka’ | 7-6 | 12.6 | Resistant |
| 4 | ‘Zimoyarka’ | 7-6 | 9.4 | Resistant |
| 5 | ‘Zolotokolosa’ | 8 | 4.7 | Highly Resistant |
| 6 | ‘Kievskaya-Ostistaya’ | 7-6 | 8.4 | Resistant |
| 7 | ‘Lasunya’ | 7-6 | 7.3 | Resistant |
| 8 | ‘Natalka’ | 8 | 5.0 | Highly Resistant |
| 9 | ‘Niva-Kievshchiny’ | 7-6 | 11.2 | Resistant |
| 10 | ‘Novokiivska’ | 7-6 | 9.4 | Resistant |
| 11 | ‘Pereyaslavka’ | 7-6 | 8.2 | Resistant |
| 12 | ‘Pivna’ | 7-6 | 12.6 | Resistant |
| 13 | ‘Podolyanka’ | 7-6 | 8.5 | Resistant |
| 14 | ‘Smuglyanka’ | 8 | 3.4 | Highly Resistant |
| 15 | ‘Sonechko’ | 7-6 | 10.0 | Resistant |
| 16 | ‘Favoritka’ | 8 | 4.3 | highly Resistant |
| 17 | ‘Khutoryanka’ | 5 | 16.7 | Moderately susceptible |
| 18 | ‘Yatran 60’ | 7-6 | 10.5 | Resistant |
| Cultivars from Remeslo Institute of Wheat of NAAS of Ukraine | ||||
| 19 | ‘Krizhinka’ | 7-6 | 9.6 | Resistant |
| 20 | ‘Mironovskaya-808’ | 7-6 | 12.2 | Resistant |
| 21 | ‘Mironovskaya-30’ | 5 | 24.2 | Moderately susceptible |
| Cultivars of Plant Breeding and Genetics Institute – National Center of Seed and Cultivar Investigation- of NAAS of Ukraine | ||||
| 22 | ‘Bilyava’ | 7-6 | 7.2 | Resistant |
| 23 | ‘Oksana’ | 7-6 | 6.3 | Resistant |
| 24 | ‘Panna’ | 7-6 | 7.8 | Resistant |
| Cultivar of National Scientific Centre “Institute of Agriculture of the National Academy of Agrarian Sciences of Ukraine” | ||||
| 25 | ‘Nedra’ | 7-6 | 9.0 | Resistant |
| Cultivars from the “Roden10” farm | ||||
| 26 | ‘Torchynska’ | 7-6 | 7.4 | Resistant |
| Foreign cultivars | ||||
| 27 | ‘Aranka’ | 7-6 | 5.6 | Resistant |
| 28 | ‘Glenlea’ | 7-6 | 4.3 | Resistant |
| 29 | ‘Granny’ | 7-6 | 5.8 | Resistant |
| 30 | ‘Triso’ | 7-6 | 6.9 | Resistant |
| 31 | ‘Tybalt’ | 7-6 | 6.2 | Resistant |
| 32 | ‘Federer’ | 7-6 | 6.1 | Resistant |
The study of wheat cultivars using molecular markers revealed that several varieties contained the gene Lr34/Yr18/Sr57/Pm38/Bdv1, which confers resistance to leaf rust. It was important to evaluate the effect of this gene on the intensity of infestation of wheat samples by leaf rust in field trials. Using the marker cssfr5, the varieties under examination were divided into two groups: those containing the resistance-associated allele Lr34(+), and those containing susceptibility-associated allele Lr34(-). Eight of the test varieties had the Lr34(+) allele, while the Lr34(-) allele was detected in most varieties.
According to the statistical analysis conducted, the correlation between resistance/lack of resistance of varieties to leaf rust in the field and the allelic composition of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene was insignificant. The level of significance of differences p> 0,05; Ffact = 1.519903 <Fteor = 3.369016; p = 0.237539. The average values of lesion intensity were: 8.65 ± 4.62 in varieties which did not have the Lr34(+) allele and 9.31 ± 2.64 in varieties that contained the Lr34(+) allele. Thus, no significant effect of the presence of the Lr34(+) allele on the manifestation of resistance to leaf rust in the varieties studied has been established.
4. DISCUSSION
The difference in the results for the microsatellite marker Xgwm295, co-dominant molecular genetic markers csLV34 and cssfr5 can be explained by the fact that the locus Xgwm295 and the locus csLV34 are not part of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene, therefore markers Xgwm295 and csLV34 are less accurate than cssfr5.
The comparative analysis has also been conducted to analyze the obtained data on the allelic state of the gene Lr34/Yr18/Sr57/Pm38/Bdv1 in Ukrainian varieties with the outcomes of previous studies. Detection of the allelic composition of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene in 181 cultivars of Ukrainian breeding was performed by Kozub et al. [31]. The authors showed that the resistance associated allele Lr34 + was identified in 42.8% of the varieties analyzed, and in varieties of PBGI breeding its frequency was 57.8%.
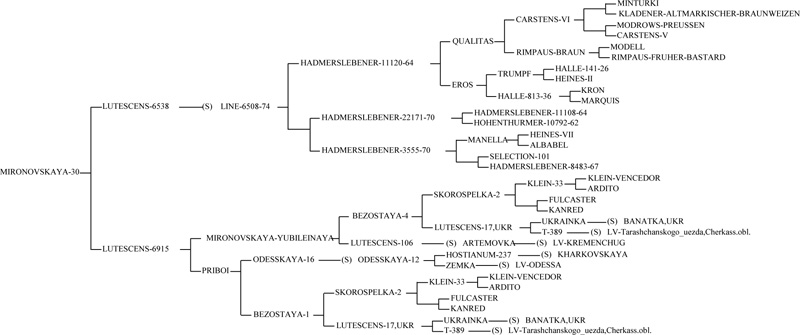
Previous research shows that the gene Lr34/Yr18/Sr57/Pm38/Bdv1 originated from an old Italian variety ‘Rieti’ [29]. Italian breeder Strampelli created the ‘Mentana’, ‘Ardito’, ‘Ballila’, and ‘Villa Glori’ varieties using the breeding line selected from ‘Rieti’, a high-yielding Dutch variety ‘Wilhelmina’ and a precocious, resistant to lodging Japanese variety ‘Akagomughi’, which had been used for crossbreeding. Thus, the ‘Frontana’ variety was obtained by using the variety ‘Mentana’, the carrier of the Lr34 + allele [29, 32]. From the variety ‘Frontana’, the gene was translocated into wheat varieties in the United States, Canada, and other countries. The donor of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene in European wheat varieties, including ‘Bezostaya 1’, is almost certainly the ‘Ardito’ variety [32, 33]. Our research results confirm the hypothesis that the Lr34/Yr18/Sr57/Pm38/Bdv1 gene was introduced into Ukrainian wheat varieties from ‘Bezostaya 1’. The cultivars ‘Panna’, ‘Mironovskaya-30’, ‘Krizhinka’, which contain the allele Lr34 (+), originate from ‘Bezostaya 1’ (Fig. 4) [32].
The high resistance of varieties with the Lr34(-) allele to the leaf rust pathogen can be explained by the presence of other resistance genes, which were beyond the scope of this work. Although no significant effect of the Lr34(+) allele on leaf rust resistance has been established, it might be appropriate to conduct a further study to elaborate a possible role of the gene on yellow rust and powdery mildew as well as spot blotch resistance (the latter one has been reported in the literature [34]) under the field conditions in Ukraine.
The results of our research are in good agreement with the previously published studies of the allelic state of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene [1, 7, 25]. However, for some varieties, the results obtained in this research were different, for instance, the varieties ‘Nedra’ and ‘Podolyanka’ were previously reported to carry the susceptible allele of the gene [5, 31] while, according to our results, they carry the allele of resistance (Table 3). As for the ‘Pivna’ variety, it was previously found that the variety was polymorphic [7, 31] while in this study, only the sensitivity allele was detected (Table 3). Such differences may be due to the heterogeneity of the seed material of wheat varieties of Ukrainian breeding used for different studies. Some varieties of Ukrainian and foreign breeding were analyzed using molecular markers of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene for the first time in this study. The results of our field research confirm the findings previously obtained: the allelic state of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene did not correlate with the field resistance of wheat varieties of Ukrainian origin as it has already has been shown by the independent trials under different conditions [34]. It can be summarized that although resistance-associated allele of the gene is widespread among wheat cultivars of Ukrainian origin its potential has not been yet fully exploited due to lack of appropriate tools (that is, molecular markers). Therefore the development of the breeding lines with a combination of the resistance-associated allele of Lr34/Yr18/Sr57/Pm38/Bdv1 and other leaf rust resistance genes may be recommended as a follow-up to this study.
CONCLUSION
DNA markers csLV34, Xgwm295 and cssfr5 were used to evaluate alleles of the Lr34/Yr18/Sr57/Pm38/Bdv1 gene in Ukrainian and foreign bread wheat cultivars. In the sample, two alleles of the csLV34 marker have been detected: csLV34a and csLV34b; three alleles for the microsatellite marker Xgwm295: 250 bp, 254 bp and 256 bp and finally, two alleles, Lr34(+) and Lr34(-), of the gene-localized marker cssfr5. The coincidence between the results obtained with the markers cssfr5 and csLV34 was 84.5%. According to our research, eight wheat samples contained the Lr34(+) allele associated with leaf rust resistance which is 25% of the total number of wheat varieties studied. Our results have also confirmed the hypothesis that the source of the gene Lr34/Yr18/Sr57/Pm38/Bdv1 in the varieties of Ukrainian breeding is ‘Bezostaya 1’.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
Dr.Yaroslav Blume is editor in chief of The Open Agriculture Journal.
ACKNOWLEDGEMENTS
This work was supported by Projects of the National Academy of Sciences of Ukraine “Introduction of molecular genetic markers of resistance to highly virulent yellow rust pathotypes of Asian origin into the selection process of wheat” (Nº0117U002730, 2017), “Creation of a molecular genetic platform for marker-assistance selection (Nº 0119U100597, 2019–2021)”.


