All published articles of this journal are available on ScienceDirect.
Molecular Characterization using Directed Amplification of Minisatellite-region DNA (DAMD) Marker in Ficus Sycomorus L. (Moraceae)
Abstract
Background:
Ficus sycomorus L. species exhibited great importance with various applications in pharmacology and medicine studies. However, little attention has been given to its molecular characterization.
Objective:
The study aimed to assess DNA genetic diversity among 16 genotypes of F. sycomorus L. species.
Methods:
Directed Amplification of Minisatellite-region DNA (DAMD) marker has been employed to investigate the genetic relationship among the studied genotypes of F. sycomorus L. species based on the estimated Percent Disagreement Values (PDV).
Results:
Twenty-four DAMD primers produced 194 bands, of which, 145 (74.742%) were polymorphic with Polymorphic Information Content (PIC) average of 0.219. DAMD-PCR application highlighted 12 unique markers characteristic for some studied genotypes. Cluster analysis showed that the studied F. sycomorus L. genotypes were split into two main distinguished clusters, each one was considered as a subspecies. In this respect, F. sycomorus14 and F. sycomorus15 genotypes were considered as subspecies too far from the second one containing the remaining genotypes.
Conclusion:
The DAMD assay successfully highlighted genetic diversity within F. sycomorus species. More accurate molecular markers are required to confirm the current data.
1. INTRODUCTION
Ficus sycomorus L. (Moraceae) species is characterized by its geographic diversity due to its adaptation to different environmental conditions. Assessment of genetic diversity of F. sycomorus L., as in the case of other fruit trees, is important not only in evolutionary studies, but also in the breeding and conservation programs of germplasm. It has been demonstrated that the existence of 400 monoecious and 800 gynodioecious species belonged to the Ficus genus [1, 2]. Overall, F. sycomorus L. species taxonomically is classified as a subgenus of the fig, belonging to the Moraceae family.
It was originated in Ethiopia and Central Africa, and planted since antiquity in Egypt, Palestine, Lebanon and Syria. Since a long time ago, Mouterde [3] reported that this species is at risk due to urban development, with little occurrence of this species in Sida and Syrian littoral zones.
This study is part of a series of studies currently underway in Syria and around the world under the project of biodiversity and the inventory of genetic resources, especially those neglected and marginalized to benefit food, medicine and genetic improvement programs to combat diseases, insects and abiotic stresses and a part of the efforts being made to expand their cultivation. F. sycomorus L. species displayed an important role as a coastal plant, genetically resource tolerant to salinity, as a food (their fruit) and medical treatment [2, 4].
Many DNA-PCR based markers are successfully employed worldwide for investigation of DNA genetic diversity in plants kingdom. Of the available techniques, Directed Amplification of Minisatellite-region DNA (DAMD) technique has been developed by Heath et al. [5] and revealed polymorphism due to minisatellites. Since this technique is carried out at higher PCR-stringencies, the patterns produced have a greater reproducibility than Random Amplified Polymorphic DNA (RAPD) [6]. It has been documented that minisatellites called as the Variable Number of Tandem Repeats (VNTR), are tandem repeats of 10-60 bp long DNA sequence motif and are widely distributed throughout the eukaryote genome. The variation in these repeats is proved to be the source of the polymorphism in several organisms [7].
It has been demonstrated that these minisatellites and DNA sequences flanking them are involved in inversions, which result in their distribution on both the strands in opposite orientations. This makes PCR possible using a minisatellite core sequence as a single primer. Moreover, these minisatellites have characteristics of inheritance in Mendelian fashion and extensive genome coverage [5].
A DAMD marker has been widely used for genetic characterization of many plant species including Oryza [8], Morus spp [9], Panax spp [10], Shorea curtisii [11], Capsicum [12], Convolvulus [13], Astragalus rhizanthus complex [14], common bean (Phaseolus vulgaris L.) [15], cucumber (Cucumis sativus L.) [16], grapevine [17], mango [18], Citrus [19], chickpea [20], Jatropha [21], Flavoparmelia caperata L [22], Dianthus L [23]. and recently, in Citrus [24] and Sechium edule (Jacq.) Sw [25].
The utility of DAMD marker, however, has not been widely reported in F. sycomorus L. germplasm characteriz ation. Thereby, the current study aimed to assess the phylogenetic relationship among F. Sycom genotypes (scattered in various areas of the Syrian coast) along with the precise identification of the subspecies belonging to the F. sycomorus species, especially since there is no precise inventory of the number of subspecies of this species in Syria and to find specific molecular markers of this species using the DAMD assay.
2. MATERIAL AND METHODS
2.1. Plant Materials
Sixteen F. sycomorus leaves samples were collected from the coastal regions of Syria (Table 1). Five to ten leaves from each genotype were bulked as one sample for DNA extraction. Table 1 shows characteristic collection sites in terms of the original site, altitude and annual rainfall. Sample collection has been performed in autumn as previously reported by Saleh [2].
2.2. DNA Extraction
Total genomic DNA of each genotype has been performed by a CTAB (cetyltrimethylammonium bromide) protocol as described by Doyle and Doyle [26]. The concentration of DNA was measured by the DNA fluorimeter and kept at –80°C until use.
2.3. DAMD-PCR Assay
Twenty-four DAMD primers were tested as previously reported in other species [9, 14, 17]. DAMD-PCR amplific ation reaction was carried out in 25 μl reaction volume containing 1X PCR buffer, 2 mM MgCl2, 0.25 mM dNTPs, 25 pmol primer, 1.5 unit of Taq DNA polymerase and 50 ng template DNA. PCR amplification was performed in a T-gradient thermal cycler (Bio-Rad; T-Gradient). It was programmed to 35 cycles after an initial denaturation cycle for 4 min at 94ºC. Each cycle consisted of a denaturation step for 1 min at 94ºC, an annealing step for 2 min at Tm varying according to each examined primer (Table 2), and an extension step at 72ºC for 2 min, followed by extension cycle for 7 min at 72ºC in the final cycle. PCR products were then separated on a 1.8% ethidium bromide-stained agarose gel (Bio-Rad) in 0.5X TBE buffer. Electrophoresis was performed for 2 h at 100 V and visualized with a UV transilluminator. A standard VC 100bp Plus DNA Ladder (Vivantis) was used to estimate the molecular weight of DAMD-PCR amplification products.
| Genotype Code | Original Site | Altitude (m) | Annual Rainfall (mm) | References |
|---|---|---|---|---|
| F. sycomor 1 | Lattakia | 4.5 | 650-700 | [2] |
| F. sycomor 2 | Lattakia | 6.5 | 650-700 | Current study |
| F. sycomor 3 | Lattakia | 6.3 | 650-700 | [2] |
| F. sycomor 4 | Lattakia | 6.5 | 650-700 | Current study |
| F. sycomor 5 | Lattakia | 5 | 650-700 | Current study |
| F. sycomor 6 | Lattakia | 5 | 650-700 | Current study |
| F. sycomor 7 | Lattakia | 5 | 650-700 | Current study |
| F. sycomor 8 | Lattakia | 5 | 650-700 | Current study |
| F. sycomor 9 | Lattakia | 4 | 650-700 | Current study |
| F. sycomor 10 | Lattakia | 6.3 | 650-700 | [2] |
| F. sycomor 11 | Lattakia | 5 | 650-700 | Current study |
| F. sycomor 12 | Jableh | 11.8 | 650-700 | [2] |
| F. sycomor 13 | Banyas | 10 | 700-750 | [2] |
| F. sycomor 14 | Banyas | 220 | ~850 | [2] |
| F. sycomor 15 | Banyas | 12 | 700-750 | Current study |
| F. sycomor 16 | Banyas | 250 | ~850 | [2] |
| Primer Number | Primer Name | Primer Sequence 5'-3' | Source Species | Ta (°C) | References |
|---|---|---|---|---|---|
| 1 | URP1F | ATCCAAGGTCCGAGACAACC | Rice | 55 | [16] |
| 2 | URP2R | CCCAGCAACTGATCGCACAC | Rice | 61 | [16] |
| 3 | URP4R | AGGACTCGATAACAGGCTCC | Rice | 58 | [16] |
| 4 | URP9F | ATGTGTGCGATCAGTTGCTG | Rice | 60 | [16] |
| 5 | URP25F | GATGTGTTCTTGGAGCCTGT | Rice | 58 | [16] |
| 6 | HVR(-) | CCTCCTCCCTCCT | Rice | 50 | [16] |
| 7 | OGRB01 | AGGGCTGGAGGAGGGC | Rice | 55 | [16] |
| 8 | FVIIex8C | CCTGTGTGTGTGCAT | Human | 47 | [16] |
| 9 | HBV3 | GGTGAAGCACAGGTG | Human | 53 | [16] |
| 10 | HBV5 | GGTGTAGAGAGGGGT | Human | 56 | [16] |
| 11 | YNZ22 | CTCTGGGTGTGGTGC | Human | 56 | [16] |
| 12 | 14C2 | GGCAGGATTGAAGC | Human | 53 | [16] |
| 13 | 33.6 | GGAGGTGGGCA | Human | 52 | [16] |
| 14 | 6.2H(+) | AGGAGGAGGGGAAGG | Human | 56 | [16] |
| 15 | M13 | GAGGGTGGCGGCTCT | Phage M13 | 57 | [16] |
| 16 | HBVb | GGTGTAGAGAGAGGGGT | Rice | 52 | [9] |
| 17 | HVRc | CCTCCTCCCTCCT | Rice | 52 | [9] |
| 18 | URP2F | GTGTGCGATCAGTTGCTGGG | Rice | 50 | [17] |
| 19 | URP6R | GGCAAGCTGGTGGGAGGTAC | Rice | 50 | [17] |
| 20 | URP13R | TACATCGCAAGTGACACAGG | Rice | 50 | [17] |
| 21 | URP17R | AATGTGGGCAAGCTGGTGGT | Rice | 50 | [17] |
| 22 | M13 | GAGGGTGGCGGTTCCT | Rice | 55 | [14] |
| 23 | HVA | AGGATGGAAAGGAGGC | Rice | 55 | [14] |
| 24 | HVY | GCCTTTCCCGAG | Rice | 55 | [14] |
2.4. DAMD Analysis
The presence or absence of each size class was scored as 1 or 0, respectively. The Percent Disagreement Values (PDV) found were used to generate a matrix via the Unweighted Pair Group Mean Arithmetic average (UPGMA) using Statistica program [27]. Then the previous matrix was used to estimate genetic similarity [28]. Polymorphic Information Content (PIC) value was estimated for each DAMD primer according to the formula:
| PIC = 1 ‒ Σ(Pij)2 |
Where Pij is the frequency of the ith pattern revealed by the jth primer summed across all the patterns revealed by the primers [29].
3. RESULTS
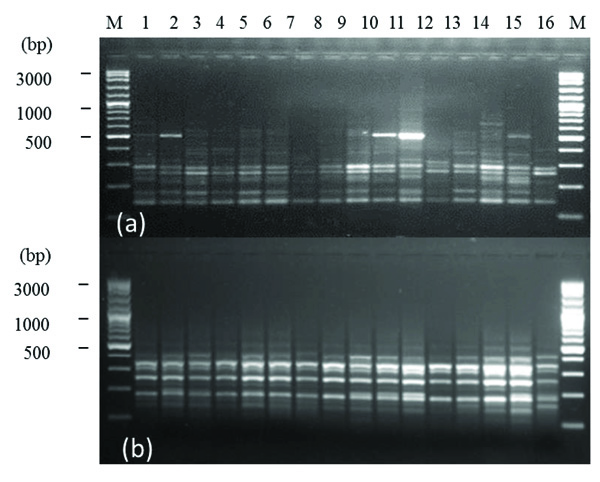
The size of DAMD-PCR amplification products ranged between 100-1500 bp. Fig. (1) shows polymorphic patterns within F. sycomorus L. species produced by 14C2 (a) and URP17R (b) DAMD primers.
The DAMD marker applied in the molecular study within F. sycomorus L. species, produced total bands number (TB) of 194 bands, of which, 145 were polymorphic (PB) representing percentage polymorphism (P%) of 74.742% (Table 3). Data showed that TB ranged between 4 [URP1F & 6.2h(+)] and 19 (URP4R) with a mean average of 8.083 bands/ primer. Whereas, PB ranged between 2 [URP9F, HVR(-), 6.2H(+) and URP6R] and 19 (URP4R) with a mean average of 6.042 bands/ primer (Table 3). As for estimated PIC, it ranged between 0.098 (URP6R) and 0.369 (M13) with a mean average of 0.219 (Table 3).
The DAMD assay successfully produced twelve unique markers, characterized F. sycomorus genotypes (Table 4).
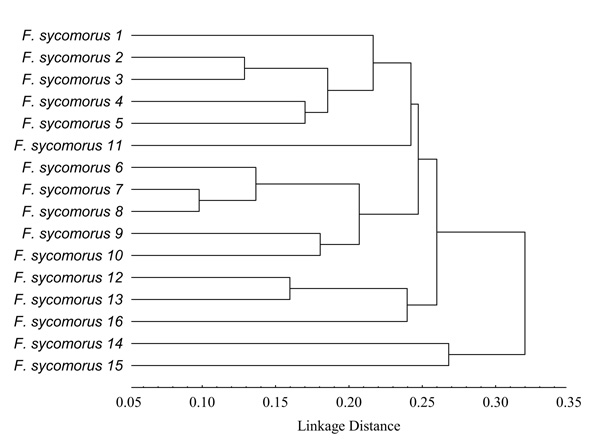
Based on UPGMA, Fig. (2) shows that the studied F. sycomorus genotypes were clustered into two main clusters. The first cluster contained F. sycomorus14 and F. sycomorus15 genotypes, whereas, the second one contained three subclusters. The first subcluster contained F. sycomorus1, F. sycomorus2, F. sycomorus3, F. sycomorus4, F. sycomorus5 and F. sycomorus11 genotypes (Fig. 2), where, F. sycomorus2 and F. sycomorus3 were too closed with the PDV value of 0.13 (Table 5). Indeed, F. sycomorus4 and F. sycomorus5 were also too closed with the PDV value of 0.17. Whereas, the second subcluster contained F. sycomorus6, F. sycomorus7, F. sycomorus8, F. sycomorus9 and F. sycomorus10 genotypes, where, F. sycomorus7 and F. sycomorus8 were the closets genotypes with the lowest PDV of 0.10. Indeed, F. sycomorus9 and F. sycomorus10 were also too closed with the PDV value of 0.18, while, the third subcluster contained F. sycomorus12, F. sycomorus13 and F. sycomorus16 genotypes, where, F. sycomorus12 and F. sycomorus13 were too closed with the PDV value of 0.16 (Table 5). The DAMD marker revealed that the lowest PDV was recorded to be 0.10 between F. sycomorus6 & F. sycomorus7 and between F. sycomorus7 & F. sycomorus8 genotypes with the highest genetic similarity of 0.831 and 0.830, respectively. Whereas, the highest one was recorded to be 0.38 between F. sycomorus8 & F. sycomorus15 (with a similarity of 0.526); F. sycomorus7 & F. sycomorus15 (with a similarity of 0.523) and F. sycomorus12 & F. sycomorus15 (with a similarity of 0.500) genotypes. It worth noting that the latest genotypes were genetically too far, exhibiting a high PDV of 0.38 than the mean PDV of 0.24.
| Primer Number | Primer Name | TB | PB | P% | PIC |
|---|---|---|---|---|---|
| 1 | URP1F | 4 | 3 | 75.000 | 0.178 |
| 2 | URP2R | 11 | 11 | 100.000 | 0.363 |
| 3 | URP4R | 19 | 19 | 100.000 | 0.366 |
| 4 | URP9F | 5 | 2 | 40.000 | 0.173 |
| 5 | URP25F | 7 | 4 | 57.143 | 0.141 |
| 6 | HVR(-) | 6 | 2 | 33.333 | 0.099 |
| 7 | OGRB01 | 9 | 6 | 66.667 | 0.22 |
| 8 | FVIIex8C | 10 | 8 | 80.000 | 0.191 |
| 9 | HBV3 | 14 | 10 | 71.429 | 0.235 |
| 10 | HBV5 | 6 | 4 | 66.667 | 0.202 |
| 11 | YNZ22 | 9 | 4 | 44.444 | 0.145 |
| 12 | 14C2 | 7 | 5 | 71.429 | 0.166 |
| 13 | 33.6 | 7 | 5 | 71.429 | 0.166 |
| 14 | 6.2H(+) | 4 | 2 | 50.000 | 0.152 |
| 15 | M13 | 9 | 8 | 88.889 | 0.297 |
| 16 | HBVb | 6 | 5 | 83.333 | 0.323 |
| 17 | HVRc | 6 | 4 | 66.667 | 0.178 |
| 18 | URP2F | 7 | 5 | 71.429 | 0.246 |
| 19 | URP6R | 5 | 2 | 40.000 | 0.098 |
| 20 | URP13R | 9 | 7 | 77.778 | 0.202 |
| 21 | URP17R | 9 | 6 | 66.667 | 0.179 |
| 22 | M13 | 11 | 11 | 100.000 | 0.369 |
| 23 | HVA | 6 | 4 | 66.667 | 0.268 |
| 24 | HVY | 8 | 8 | 100.000 | 0.287 |
| Total | 194 | 145 | − | − | |
| average | 8.083 | 6.042 | 70.374 | 0.219 |
| Genotype Code | Original SiteS | Primer | Unique Positive Marker Molecular Weight (bp) |
|---|---|---|---|
| F. sycomor 1 | Lattakia | FVIIex8C | ~150 |
| F. sycomor 1 | Lattakia | 33.6 | 500 |
| F. sycomor 1 | Lattakia | URP2F | ~290 |
| F. sycomor 4 | Lattakia | URP25F | ~150 |
| F. sycomor 6 | Lattakia | URP13R | 200 |
| F. sycomor 8 | Lattakia | URP4R | ~450 |
| F. sycomor 9 | Lattakia | HVRc | 200 |
| F. sycomor 9 | Lattakia | HVY | 200 |
| F. sycomor 13 | Banyas | URP4R | ~150 |
| F. sycomor 13 | Banyas | FVIIex8C | ~1100 |
| F. sycomor 14 | Banyas | URP4R | 900 |
| F. sycomor 14 | Banyas | HBV3 | 1500 |
| F. sycomor 15 | Banyas | 33.6 | 1000 |
| F. sycomor 15 | Banyas | URP2F | 100 |
| F. sycomor 15 | Banyas | URP6R | 800 |
| F. sycomor 15 | Banyas | URP13R | ~450 |
| F. sycomor 15 | Banyas | M13 | ~150 |
| F. sycomor 16 | Banyas | URP1F | ~190 |
| F. sycomor 16 | Banyas | 6.2H(+) | 200 |
| Genotype | F. syc1 | F. syc2 | F. syc3 | F. syc4 | F. syc5 | F. syc6 | F. syc7 | F. syc8 | F. syc9 | F. syc10 | F. syc11 | F. syc12 | F. syc13 | F. syc14 | F. syc15 | F. syc16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F. syc1 | 0.00 | |||||||||||||||
| F. syc2 | 0.19 | 0.00 | ||||||||||||||
| F. syc3 | 0.23 | 0.13 | 0.00 | |||||||||||||
| F. syc4 | 0.22 | 0.17 | 0.21 | 0.00 | ||||||||||||
| F. syc5 | 0.23 | 0.19 | 0.18 | 0.17 | 0.00 | |||||||||||
| F. syc6 | 0.29 | 0.21 | 0.27 | 0.21 | 0.21 | 0.00 | ||||||||||
| F. syc7 | 0.29 | 0.25 | 0.28 | 0.22 | 0.24 | 0.10 | 0.00 | |||||||||
| F. syc8 | 0.27 | 0.23 | 0.27 | 0.19 | 0.24 | 0.17 | 0.10 | 0.00 | ||||||||
| F. syc9 | 0.29 | 0.24 | 0.24 | 0.25 | 0.24 | 0.22 | 0.22 | 0.16 | 0.00 | |||||||
| F. syc10 | 0.24 | 0.22 | 0.24 | 0.22 | 0.24 | 0.23 | 0.22 | 0.20 | 0.18 | 0.00 | ||||||
| F. syc11 | 0.23 | 0.25 | 0.26 | 0.23 | 0.24 | 0.32 | 0.28 | 0.26 | 0.26 | 0.21 | 0.00 | |||||
| F. syc12 | 0.25 | 0.29 | 0.29 | 0.27 | 0.29 | 0.30 | 0.26 | 0.25 | 0.28 | 0.18 | 0.22 | 0.00 | ||||
| F. syc13 | 0.27 | 0.24 | 0.31 | 0.24 | 0.28 | 0.28 | 0.27 | 0.24 | 0.30 | 0.19 | 0.27 | 0.16 | 0.00 | |||
| F. syc14 | 0.31 | 0.25 | 0.26 | 0.27 | 0.28 | 0.32 | 0.30 | 0.32 | 0.35 | 0.27 | 0.28 | 0.31 | 0.25 | 0.00 | ||
| F. syc15 | 0.37 | 0.31 | 0.37 | 0.35 | 0.36 | 0.32 | 0.38 | 0.38 | 0.35 | 0.34 | 0.34 | 0.38 | 0.30 | 0.27 | 0.00 | |
| F. syc16 | 0.26 | 0.23 | 0.26 | 0.25 | 0.29 | 0.28 | 0.23 | 0.23 | 0.26 | 0.24 | 0.27 | 0.23 | 0.25 | 0.29 | 0.35 | 0.00 |
DISCUSSION AND CONCLUSION
DNA genetic diversity among 16 genotypes of F. sycomorus L. species has been assessed using 24 DAMD primers. Data showed that TB ranged between 4-19 bands with a mean average of 8.083 bands/ primer, whereas, PB ranged between 2-19 bands with a mean average of 6.042 bands/ primer. Another investigation by Saleh [2] reported DNA genetic diversity among 10 F. sycomorus genotypes using RAPD and Inter-retrotransposon Amplified Polymorphism (IRAP) markers. The previous study revealed that the RAPD marker produced TB ranging between 5-21 bands with an average of 9.778 bands/primer and PB ranging between 2-16 bands with an average of 7 bands/primer. Whereas, in IRAP marker case, TB ranged between 7-17 bands with an average of 11.125 bands/primer and PB ranged between 3-15 bands with an average of 9.438 bands/primer.
Previously, Zhou et al. [8] reported high levels of variation between different species and little variation between different cultivars of O. sativa using DAMD. Whereas, Ince et al. [12] reported that TB varied from 1 to 12 fragments combined with a total of 38 accession-specific DNA markers in Capsicum using 22 DAMD primers.
Moreover, Seyedimoradi et al. [17] reported a DAMD marker for characterizing 21 grapevine accessions from Iran. The previous study revealed that P% average was recorded to be 66% with a PIC average value of 0.44. Indeed, genetic dissimilarity of genotypes ranged from 0.12 to 0.67. Srivastava et al. [18] applied 4 DAMD primers for genetic diversity studying among 46 mango varieties. They reported 110 (with P%=87.3%) fragments with a PIC value of 0.245. In addition, Kumar and Nair [19] applied 4 DAMD primers to study phylo genetic relationships among 50 wild and cultivated accessions of 19 Indian Citrus genotypes. They reported a total of 45 bands, of which 35 (78%) were polymorphic with a mean genetic similarity of 0.68, indicating moderate genetic divergence among the studied accessions. Moreover, Pakseresht et al. [20] reported a DAMD marker for studying phylogenetic relationships of 40 landraces chickpea genotypes in Iran. The previous study revealed the PIC average 0.232, whereas, Murty et al. [21] applied 4 DAMD primers for studying genetic diversity in 19 Jatropha accessions. The previous study revealed that P% average was 91.02% with a mean PIC value 0.873. Indeed, Ince and karaca [23] applied 22 DAMD primers to study genetic diversity in Dianthus L. genus. The previous study revealed that TB varied between 36-168 bands and PB varied between 19-120 bands with P% ranging between 37.3-100% and PIC values ranging between 0.120-0.341.
Recently, Pinar et al. [24] applied 20 DAMD primers for Citrus molecular characterization. The previous study revealed that DAMD marker gave 296 total bands, of which, 296 (100%) were polymorphic with an average of 14.8 bands/primer. Whereas, Jain et al. [25] applied 12 DAMD primers for Sechium edule (Jacq.) Sw. molecular study. They reported a total of 97 bands (average of 8.033 bands/primer) of which 92 (average of 7.667 bands/primer) were polymorphic with P% of 94.845%.
The current study revealed that DAMD assay produced 194 bands, of which, 145 (74.742%) were polymorphic with a PIC average value of 0.219. Saleh [2] however, reported the molecular characterization of 10 F. sycomorus genotypes using 36 RAPD and 22 IRAP primers. The previous study revealed that RAPD assay produced 352 distinguished bands, of which 252 (71.59%) were polymorphic. As for IRAP assay, there were 178 bands, of which, 151 (84.83%) were polymorphic. Moreover, the mean PIC value was recorded to be 0.215 and 0.299 for RAPD and IRAP assays, respectively.
It worth noting that our data presented herein were comparable with that reported by Saleh [2]. Since, the effectiveness of a molecular marker for detection of DNA genetic diversity is based on P% and PIC values resulting from the application of the given marker, in this respect, in our case study, the DAMD marker was more effective than RAPD by showing the highest P% (74.742%) and PIC (0.219) values compared to RAPD with the lowest P% (71.59%) and PIC (0.215) values. This observation was in agreement with Karaca and Ince [6] who reported that the DAMD marker was more effective than the RAPD one. While, the DAMD marker was less effective than the IRAP one in the study by Saleh [2], where, IRAP marker exhibited the highest P% (84.83%) and PIC (0.299) values than that those reported in the current study.
Overall, based on data previously reported by Saleh [2] and herein regarding P% and PIC values, we could suggest that moderate genetic divergence was observed among the tested F. sycomorus genotypes.
From data presented herein, the current study could suggest that the 16 studied F. sycomorus genotypes belonged to two separate distinguished subspecies; the first subspecies contained F. sycomorus14 and F. sycomorus15 genotypes. Whereas, the second subspecies contained the remaining studied genotypes.
Overall, the DNA genetic diversity detected among the studied F. sycomorus genotypes was not observed to be related to their geographical distribution, but could be related to their common origin as previously reported by Saleh [2] who stated similar observation within F. sycomorus species. This finding was coherent with those reported by Machado et al. [30], showing the evolution of Ficus monoecious origin into gynodioecious types. Other studies, however, reported similar findings related to Tunisian fig based on the RAPD assay application [31].
In conclusion, data presented herein using the DAMD maker and in the previous study [2] using RAPD and IRAP markers, highlighted moderate genetic divergence among the studied F. sycomorus genotypes. Overall, the current study suggests that the studied F. sycomorus genotypes were split into two main subspecies. Indeed, DAMD-PCR assay successfully highlighted 12 unique markers characterized some studied genotypes. Thereby, due to the importance of F. sycomorus as a coastal species and to confirm the actual results obtained regarding the mentioned species for DNA genetic diversity; further research works like e.g. Simple Sequence Repeats (SSRs) are required to confirm the number of sub species to which F. sycomorus trees growing in Syria belong; especially, there is no precise inventory of the number of subspecies of this species in Syria.
LIST OF ABBREVIATIONS
| CTAB | = Cetyltrimethylammonium Bromide |
| DAMD | = Directed Amplification of Minisatellite-region DNA |
| IRAP | = Inter-retrotransposon Amplified Polymorphism |
| P% | = Percentage Polymorphism |
| PB | = Polymorphic Bands |
| PIC | = Polymorphic Information Content |
| PDV | = Percent Disagreement Values |
| RAPD | = Random Amplified Polymorphic DNA |
| SSRs | = Simple Sequence Repeats |
| TB | = Total Bands |
| UPGMA | = Unweighted Pair Group Mean Arithmetic Average |
| VNTR | = Variable Number of Tandem Repeats |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
I thank Dr. I. Othman (Director General of AECS) and Dr. N. Mirali (Head of Molecular Biology and Biotechnology Department in AECS) for their support, and also the Plant Biotechnology group for technical assistance.


