All published articles of this journal are available on ScienceDirect.
Genetic Characterization and Growth Promotion Mechanisms of Burkholderia vietnamiensis Isolated from Rice Cultivars in Peru
Abstract
Introduction/Background
Burkholderia is a versatile bacterial genus, and from a biotechnological point of view, it is a source of diverse secondary metabolites with enormous application potential, especially in agriculture.
Objective
This study aimed to isolate diazotrophic Burkholderia bacteria-associated rice roots and study the genetic and PGPR diversity among strains and the effect of their inoculation in two rice cultivars.
Methods
Strains were isolated using nitrogen-free semisolid media and tested by specific amplification of the recA gene. The production of auxins, siderophores, phosphate solubilization, and antagonism against phytopathogenic fungi was evaluated, and finally, their inoculation into two rice varieties was also examined.
Results
Only 5.13% of the isolated strains were positive for the amplification of the recA gene with Burkholderia-specific primers. Sequence analysis showed high similarity with Burkholderia vietnamiensis. These strains produced auxins in tryptophan-supplemented broth (up to 13.98 µg mL-1), siderophores (up to 139.52%), phosphate solubilization (up to 15.99 mg PO4 mL-1), as well as antibiotic and antagonistic capacities against five pathogenic fungi of rice. These strains increased the vigor index in two rice cultivars compared to the non-inoculated or non-fertilized treatment. The antibiotic and antifungal activities of B. vietnamiensis strains against two pathogenic fungi, Nakataea sigmoidea and Nigrospora oryzae, are described for the first time.
Conclusion
Due to the taxonomic affinity of our strains within the Burkholderia cepacia complex, their direct use in agriculture is not recommended; however, further research is required to exploit their biotechnological potential for the synthesis of useful metabolites.
1. INTRODUCTION
Burkholderia sensu lato (s.l.) [1] is a group of Gram-negative bacteria that are ubiquitous in different ecological niches and belong to the subphylum β-proteobacteria. Currently, by conducting phylogenetic analyses of conserved genes and comparative genomics, Burkholderia s.l. is classified into 7 new genera: Burkholderia sensu stricto (s.s.), Caballeronia [2], Paraburkholderia [3], Robbsia [4], Mycetohabitans, Trinickia [5], and Pararobbsia [6]. Within the genus Burkholderia, the Burkholderia cepacia complex (Bcc) comprises a group of species from different ecological niches and metabolic versatility [7]. It is characterized by mobility, strictly aerobic oxygen requirement, and great metabolic versatility due to the diversity of ecological niches it occupies [8]. It includes phytobeneficial species that promote plant growth, such as B. ambifaria [9], B. catarinensis [10], B. orbicola [11], and B. vietnamiensis [12], as antagonists of phytopathogenic fungi and producers of metabolites of biotechnological interest [13, 14]. Burkholderia isolates have enormous biotechnological potential, as many strains are producers of hydrolytic enzymes and bioactive substances that promote plant growth and health and degrade several recalcitrant pollutants [15-17]. Some members of the Bcc have also been reported as opportunistic pathogens in humans and animals [16, 18], preventing their use as bioinoculants in the field. Plants have co-evolved with various microorganisms, allowing them to survive in hostile environmental conditions. This association between plants and microorganisms takes place at the level of the rhizosphere, endosphere, and/or phyllosphere [19]. The relationships between plants and Burkholderia sensu stricto can be mutualistic or pathogenic. Mutualistic or phytobeneficial relationships are usually associated at the rhizosphere level [4, 20, 21] or at the endosphere level [22-24]. Among the phytobeneficial Burkholderia sensu stricto, B. vietnamiensis [25] is considered a diazotroph characterized by a mutualistic relationship with Oryza sativa and considered a model bacterium for growth promotion in rice [12, 26]. Several studies have reported that B. vietnamiensis strains promote plant growth through direct mechanisms, such as the production of phytohormones, siderophores, 1-aminocyclopropane-1-carboxylate (ACC) deaminase, and nitrogen fixation, and indirect promotion mechanisms, such as biocontrol associated with the production of metabolites [27, 28]. B. vietnamiensis has been reported as a diazotroph associated with Ficus tikoua [28], Ipomoea batatas [29], and Populus trichocarpa [30] and is frequently found in rice [31-36]. The interactions between B. vietnamiensis and rice are very competent and persistent in the rhizosphere, Synthesizing a large number of secondary metabolites and expressing direct and indirect growth mechanisms [12]. Recently, in a study [37], it was demonstrated that B. vietnamiensis could inhibit root-knot nematode on watermelon by modifying the rhizosphere microbial community. There is evidence for the growth-promoting potential of B. vietnamiensis in rice [20, 31, 34]. Recently, it has been reported that B. vietnamiensis improved the growth of rice seedlings, enhanced root development, and increased nitrogen mobilisation and assimilation [20]. Unfortunately, these phytobeneficial species isolated from the rhizosphere and endosphere of plants could not be used as inoculants on a commercial scale due to a moratorium imposed in the 1990s on the grounds that they could pose a health risk [15]. However, in recent years, there has been an increase in the number of new Burkholderia species that are beneficial to plants but are not isolated from clinical sources. For this reason, there is an urgent need to develop new strategies to reduce the use of nitrogenous fertilizers, which can be addressed through the study of phytobeneficial Burkholderia associated with rice cultivation in the San Martin region. It is important to study the genetic diversity among Burkholderia species and explore their taxonomy, especially to distinguish strains with phytobeneficial potential from those with opportunistic pathogenic capacity. In addition, considering the diversity and identity of phytobeneficial Burkholderia strains, we selected strains that enable better outcomes at a level of genomic resolution. The aims of this work were: 1) to isolate and select bacteria of the genus Burkholderia sensu stricto associated with rice roots grown in four valleys of the San Martín region, using specific molecular markers, 2) to study genetic diversity and the phylogenetic relationship among Burkholderia sensu stricto strains, and 3) to evaluate the direct and indirect growth promotion mechanisms of B. vietnamiensis strains and their response to inoculation in two local rice cultivars.
2. MATERIALS AND METHODS
2.1. Collection of Rice Samples
Root samples of different rice varieties were collected from the Bajo Mayo, Alto Mayo, Alto Huallaga, and Central Huallaga valleys in the San Martín-Perú region. Root samples were obtained from plants at the tillering stage, according to the procedure described by Ji et al. [38]. Root samples were processed, coded, and transported in falcon tubes (50 mL) at 4°C. Georeferencing data and cultivation history were recorded for each sample.
2.2. Isolation of Diazotrophic Bacteria Associated with Rice Roots
Diazotrophic bacteria were isolated from roots according to the method proposed by Baldani et al. [39]. Roots were washed with copious amounts of water to remove soil remnants and immersed successively in 70% ethanol solution for 3 min, sodium hypochlorite solution (2.5%) for 5 min, 70% ethanol solution for 30 s, and rinsed five times with sterile distilled water [39]. To confirm the success of the disinfection process, 100 µL of sterile distilled water from the last rinse of each sample was streaked on the surface of plates containing Tryptone Soy Agar (TSA) and incubated at 28 °C for 3 days [35]. Only samples with no growth were considered suitable for further analysis. Root tissues were macerated with sterile physiological saline solution (0.85%), and 100 µL of them were inoculated into JMV nitrogen-free semi-solid medium [40, 41] containing (g L-1) mannitol (5.0), K2HPO4 (0.6), KH2PO4 (1.8), MgSO4 7H2O (0.2), NaCl (0.1), CaCl2.2H2O (0.2), bromothymol blue (5 g L-1 in 0.2 N KOH) (2 mL), FeEDTA (16.4 g L-1) (4 mL), micronutrient solution (2 mL), and vitamin solution (1 mL). Distilled water was added to make up 1000 mL; pH was adjusted to 5.2 ± 0.2 with KOH, and agar was added at a rate of 1.8 g L-1. The appearance of a pellicle under the culture medium after 4 days of incubation indicated the presence of diazotrophic bacteria. This bacterial film was streaked in Petri dishes containing nitrogen-free solid JMV medium (agar, 15 g L-1) and incubated at 30°C for 5 days until colonies appeared. Isolates with different morphocolonial characteristics in each sample were purified by the streaking technique in Petri dishes containing TSA until pure cultures were obtained. The preservation of the collection was carried out by cryopreservation at -20°C and -80°C, as suggested by García and Cotter [42] and Cui et al. [43].
2.3. Genomic DNA Extraction
Genomic DNA was extracted from bacterial cultures incubated overnight in Tryptone Soy Broth (TSB) (25 mL) at 28°C, shaken at 150 rpm, and the cell pellet was harvested by centrifugation at 13,000 rpm for 3 min [10]. The commercial GenEluteTM Bacterial Genomic DNA kit (Sigma Aldrich, USA) was used according to the manufacturer's instructions. The DNA concentration was measured in a NanoDrop one spectrophotometer (Thermo Scientific, USA), and the quality was verified in a 1% agarose gel, using DiamondTM Nucleic Acid Dye (Promega, USA) and visualised in a black light electrophoresis chamber (Cleaver Scientific Ltd., UK). The original DNA was stored at -20°C and -80°C.
2.4. Molecular Discrimination of Burkholderia
Strains belonging to the genus Burkholderia were selected using specific primers recA (Spilker et al. 2009). The primers recA-F (forward) 5’-AGGACGATTCATGGAAGAWAGC-3’ and recA-R (reverse) 5’-GACGCACYGAYGMRTAGAACTT-3’ amplified a specific sequence of approximately 704 bp of the recA gene. The polymerase chain reaction was performed in a Biometra thermal cycler (Analytic Jena, Germany). PCR reactions were performed using a 25 µL reaction mix containing 1.0 µL template DNA (50 ng), 12.5 µL KAPA Taq ReadyMixTM (2x) (Sigma-Aldrich, USA) (DNA polymerase, 0.05 units µL-1, 3 mM MgCl2, 400 µM each dNTP), 1.0 µL of each primer (5 pmol each forward and reverse), and 9.5 µL of PCR grade water (Sigma-Aldrich, USA). The PCR cycle was as follows: initial denaturation at 95°C for 2 minutes, 30 cycles of denaturation for 30 s at 94°C, annealing at 58°C for 30 seconds, an extension for 60 seconds at 72°C, and a final extension at 72 °C of 5 min. Two µL of each PCR product was visualized on a 1.0% agarose gel in 0.5X TAE buffer, according to Mahenthiralingam et al. [44]. Positive amplification of the fragment allowed the discrimination of strains of the genus Burkholderia. The PCR products were purified and sequenced by MACROGEN Inc. (South Korea).
2.5. BOX-PCR Genomic Profiles
Genomic profiles were obtained by amplifying genomic DNA using the BOX-A1R primer (5'-CTACGGCAAGGCGACGCTGACG-3') [45] in a Biometra Tone thermal cycler (Analytic Jena), following the cycle program described by Arone. et al. [46]. PCR products were separated on a 1.5% agarose gel running at 60 V for 8 hours and then visualised in a black light electrophoresis chamber (Cleaver Scientific Ltd., UK). The DirectLoadTM 1 kb DNA Ladder (Sigma Aldrich, USA) was used as a molecular weight marker. The genomic profiles of the individual strains generated were photographed, digitised, and converted into a binary matrix of the presence or absence of DNA bands. Cluster analysis allowed the construction of dendrograms using the free program DendroUPGMA (genomes.urv.es/UPGMA/) [47], applying the UPGMA algorithm (Unweighted Pair-Group Method with Arithmetic Mean) [48], and the Jaccard t coefficient [49], with 2% tolerance.
2.6. recA Gene Phylogeny
The recA sequences obtained were compared with those from GenBank using the BLASTN algorithm [50]. Sequences of all types of strains of the genus Burkholderia sensu stricto were obtained from the NCBI database and aligned using ClustalW [51]. Phylogenetic trees were reconstructed using a maximum likelihood (ML) approach [52] using the MEGA X software [53]. Pairwise distances were calculated using MEGA X [53] with the two-parameter Kimura model [54]. The topological robustness of the ML tree was inferred by non-parametric bootstrap analysis based on 100 pseudoreplicates.
2.7. Characterization of Growth Promotion Mechanisms
Standardised inocula of five B. vietnamiensis strains representing each different BOX-PCR group were cultured in TSB broth at 150 rpm and incubated overnight at 28°C. Cells were harvested by centrifugation at 13,000 rpm for 3 min [10] and washed at least twice with 0.85% sterile physiological saline. The cell suspension was standardised to an OD600 nm of 1.0 and used to evaluate growth promotion mechanisms.
2.7.1. Auxin Production
Auxin production was assayed by seeding 200 µL of standardised inoculum in 20 mL of TSB broth supplemented with L-tryptophan as precursor [38, 55] at increasing concentrations (0, 50, 100, 200, 400, and 600 µg mL-1). Treatments were incubated for 24 h at 26±2°C with shaking at 150 rpm. The supernatant of each treatment was obtained by centrifugation at 8000 rpm for 15 minutes at 4°C. Salkowski's reagent (150 mL of H2SO4 mixed into 250 mL of distilled water and 7.5 mL of 0.5 M FeCl3.6H2O solution) was mixed with the supernatant in equal parts (1:1) and left in the dark for 30 minutes. The absorbance of the mixture was then measured at 535 nm in a spectrophotometer (PG Instruments Ltd, UK). The concentration of IAA was estimated from a standard curve.
2.7.2. Nitrogen Fixation
Asymbiotic nitrogen fixation capacity was tested according to Ríos-Ruiz et al. [56]. The nitrogen fixation capacity was tested in a mineral medium without nitrogen (MM-N [57] (g L-1) - mannitol, 10.0 g; sucrose, 10.0 g; K2HPO4, 0.6551 g; KH2PO4, 0.15 g; FeCl3, 0.0034 g; Na2MoO4. 2 H2O, 0.0108 g; NaCl, 0.02 g; CaCl2. 2 H2O, 0.01; MgSO4. 2H2O, 0.2 g; 1.6 g (semi-solid) or 16 g (solid) agar-agar, 0.5 mL bromothymol blue solution (0.5% in 70% ethanol), pH 7.0 ± 0.2). The standardised inocula were seeded on the surface of the semi-solid MM-N at a rate of 10% (150 µL per tube). The formation of a sub-pellicle under the semi-solid MM-N culture medium and the appearance of colonies in the solid MM-N medium, as well as the turning of the bromothymol blue indicator, indicated nitrogen fixation activity.
2.7.3. Siderophores Production
The qualitative production of siderophores by B. vietnamiensis strains was detected on chrome azurol-S (CAS) agar [58]. CAS agar consisted of a mixture of a CAS staining solution (solutions 1 and 2) and a succinate medium (SM). For solution 1 (60 mL), 60.5 mg of CAS (Hi-media, India) was dissolved in 50 mL of distilled water (pH 7.0) and then mixed with 10 mL of 1 mM FeCl3 6 H2O (5.4 mg FeCl3.6 H2O) dissolved in 10 mM HCl (0.822 mL of 37% HCl and density of 1.19 in 100 mL of distilled water). Using a magnetic stirrer, solution 1 was slowly added to solution 2 (72.84 mg HDMTA (Merck, Germany) dissolved in 40 mL distilled water, pH 7.0). The resulting solution (100 mL) was autoclaved at 121°C for 20 minutes and cooled to 50°C. For the SM culture medium, 720 mL of SM broth was added to 27.45 g of PIPES (Sigma-Aldrich, USA), and the pH was reduced to 3.0 and corrected to 6.8 using a 30% KOH solution. The final mixture was made up to 810 mL with SM broth and supplemented with 15.0 g of agar-agar, then autoclaved at 121°C for 20 minutes and cooled to 60°C. The CAS agar was completed by mixing 810 mL of SM culture medium and 90 mL of CAS staining solution and then distributing the CAS agar in Petri dishes. Each strain was separately inoculated (20 µL) onto Petri dishes containing CAS agar and incubated at 28 ± 2°C for 72 hours in triplicate. The plates were scored by the colour change from blue to orange, the diameter of the siderophore production halo was evaluated, and the efficiency of siderophore production was determined using the following formula:
E%= [(halo diameter - colony diameter)/colony diameter] x 100. Samples were analysed in triplicate.
2.7.4. Phosphate Solubilization
Phosphate solubilization was evaluated qualitatively according to the method of Ramírez-Bahena et al. [59] and quantitatively according to Marra et al. [60] and Song et al. [61], both performed in agar and National Botanical Research Institute's phosphate (NBRIP) broth, respectively. The NBRIP medium had the following composition: glucose 10 gL-1, Ca3(PO4)2 5 g, (NH4)2SO4 0.1 g, MgCl2.6H2O 5 g, MgSO4.7H2O 0.25 g, and KCl, 0.2 g [62]. The pH was adjusted to 7.0 prior to autoclaving. For qualitative assessment, the NBRIP broth was supplemented with 1.5% agar (Hi-media, India), autoclaved, and served in Petri dishes. For each strain, 20 µL of standardised inoculum was inoculated in quadruplicate into a Petri dish containing NBRIP agar. The inoculated plates were incubated at 28°C, and the solubilization diameter halo (the translucent area around the colony) was measured using an electronic vernier after 15 days of incubation. The solubilisation index (SI) = halo diameter (mm)/colony diameter (mm) was determined as proposed by Marra et al. [60]. For quantitative evaluation, 200 µL of standardised inoculum was inoculated in triplicate into 20 mL of NBRIP broth and incubated at 26 ± 2°C at 150 rpm for 24 h. At the end of this period, the inoculum was discarded. The supernatant was centrifuged (13,500 rpm for 5 minutes), the pH was measured, and the amount of soluble P was quantified using the phosphomolybdate method [63]. The blank treatment consisted of the NBRIP broth without inoculation and shaking. The strains with phosphate solubilising capacity were those with a soluble P concentration higher than the blank treatment.
2.7.5. Antibiosis and Antagonism against Phytopathogenic Fungi
The methods used to study the indirect mechanisms related to the biocontrol of phytopathogenic fungi in rice in five strains of B. vietnamiensis were antagonism and antibiosis, as proposed by Castellano-Hinojosa and Bedmar [64] and Ríos-Ruiz et al. [34]. Several phytopathogenic fungi, i. e. Rhizoctonia solani (RS1), Rhizoctonia oryzae (RO1), Rhizoctonia oryzae-sativae (ROS1), Nakataea sigmoidea (NS1), and Nigrospora oryzae (NO1), were donated by the Plant Disease Diagnostic Centre, Faculty of Agrarian Sciences-Universidad Nacional de Tumbes, and were cultured on potato dextrose agar (PDA).
For antagonism assays, fungi were first grown in PDA medium and used to take 1 cm diameter agar plugs, which were placed in the center of plates containing PDA medium that had previously been independently inoculated with 100 μL of each of the strains grown in TSB. The cells were incubated for 72 hours at 25 °C, and the diameter of the inhibition zones was recorded after 7 days. The percentage of inhibition was calculated using the formula: % inhibition = [(R1-R2/R1) x 100, where R1 is the diameter of the fungal mycelium in plates not inoculated with the bacterial culture and R2 is the diameter of the fungal mycelium in plates inoculated with the bacterial culture. For the antibiosis test, the agar plugs containing the fungus were placed in the center of a plate containing PDA medium, with the strains to be inoculated in the three equidistant quadrants of the plate at a rate of 20 µL for each strain. The inhibitory effect on fungal growth was evaluated after 7 days at 25 °C, and the percentage of inhibition relative to the control (without bacteria) was evaluated as indicated above. Both assays were performed in triplicate and incubated for 48 h at 25 °C.
2.8. Evaluation of Growth Promotion in two Rice Cultivars
For the standardization of inocula, the methodologies proposed by Granada et al. [65] for germination and vigour index assays and by Wallner et al. [66] for growth promotion assays were applied. B. vietnamiensis strains were cultured in 3 mL of TSB and incubated at 28°C for 24 h at 170 rpm. The fermented broth was centrifuged at 10,000 rpm for 10 minutes at 4°C to harvest the cells. The cell pellet was washed at least twice with sterile 10 mM MgSO4 solution at 10,000 rpm for 5 minutes. The cell suspension was adjusted to an OD600 nm of 0.001 (approximately 2 x 106 CFU mL-1) for the germination and vigour index assays and to an OD600 nm of 1.0 (approximately 5 x 109 CFU mL-1) for the growth promotion assay. Finally, the cell count was verified by the drop method in Petri dishes containing TSA agar [67] in INIA-510, La Conquista (LC) and INIA 507, La Esperanza (LE), both from the National Rice Programme, El Porvenir-San Martín Experimental Station of the Instituto Nacional de Innovación Agraria (INIA). For the trials, seeds of both varieties were surface disinfected according to the methodology proposed by Ji et al. [38]. Seeds were immersed in a 5% NaOCl solution for 8 min, rinsed three times with sterile distilled water, immersed once in a 70% ethanol solution, and finally rinsed six times with sterile distilled water.
2.8.1. Germination Test and Vigor Index
To evaluate the germination percentage and the vigor index, a completely randomized experimental design was used, with a factorial arrangement of seven treatments for two rice varieties (LC and LE) and five replicates of ten seeds each. The inoculated treatments consisted of five strains of B. vietnamiensis (T1 to T5), a treatment inoculated with the control strain Priestia megaterium SMBH14-02 [56], and a blank treatment. Fifty disinfected seeds were placed in a sterile tube (50 mL) and inoculated by immersion with 25 mL of standardized inoculum per strain and 25 mL of 10 mM MgSO4 solution for the blank treatment. The treatments were incubated for two hours at room temperature under the same conditions. The inoculated seeds were sown on absorbent paper at the bottom of Magenta® boxes (200 mL) previously sterilized at 121°C for 30 minutes and moistened with 4 mL of sterile distilled water. The treatments were incubated in the dark at 25 ± 1°C for 10 days, and the following parameters were evaluated: shoot length (SL) (cm), root length (RL) (cm), germination percentage (%), and vigor index using the following formula:
Vigor index = (SL + RL) * germination (%)
2.8.2. Growth Promotion Evaluation
To evaluate growth promotion, a completely randomized experimental design was used, with a factorial arrangement of seven treatments for two rice varieties (LC and LE) and with ten replicates each. The inoculated treatments consisted of five strains of B. vietnamiensis (T1 to T5), a treatment inoculated with the control strain P. megaterium SMBH14-02 [56], and a blank treatment. The disinfected seeds were placed in Petri dishes (15 x 150 mm) containing 1% (w/v) sterile water agar and germinated at 25 ± 1°C for 3 days in the dark until 1 cm radicles were obtained. The germinated seeds were sown in previously disinfected polypropylene pots (9 x 9 cm, 250 mL) with 85 g of fertilized and sterilized greenhouse substrate (Klasmann TS 1®, Lithuania) irrigated with 55 mL of sterile water distilled to reach a capacity of 60% field. Inoculation was carried out at a rate of 1 mL of standardized inoculum per seed and 1 mL of 10 mM MgSO4 solution for the blank treatment. The treatments were watered at least twice a week with 50% Hoagland nutrient solution supplemented with 1% KNO3 to 60% field capacity through an irrigation tube installed in each pot. The seedlings were maintained under controlled conditions (80% humidity at 28°C, 16 h light: 8 h dark), and after 21 days, the following parameters were evaluated: root length (RL) (cm), shoot length (SL) (cm), total length (TL) (cm), shoot dry weight (SDW) (mg), root dry weight (RDW) (mg), total dry weight (TDW) (mg), percentage of nitrogen in the shoot (%NPA), percentage of nitrogen in the root (%NPR), and percentage of total nitrogen (%NT).
2.9. Statistical Analyses
The normality and homoscedasticity of the data were tested. Significant differences between strains and their effects on the growth promotion mechanisms in two rice varieties were studied using an analysis of variance (ANOVA) using the R command package in R studio desktop version 1.3.1093. The Tukey’s test was carried out with a significance level of 95% (P ≤ 0.05) when parametric data were available; otherwise, the Kruskal-Walli test was conducted with a significance level of 95% (P ≤ 0.05). Correlations between variables were estimated using Pearson's correlation. The difference in germination rate between the two cultivars was tested with a two-tailed t-test, and the difference in germination rate for each cultivar, whether inoculated with inocula or with MgSO4 solution, was tested with the empirical rule, where 99.7% of the values are within three standard deviations of the mean.
3. RESULTS
3.1. Isolation and Characterization of Strains
We collected 132 root samples from cultivated rice varieties in the four rice valleys of Bajo Mayo (30), Alto Mayo (57), Huallaga Central (15), and Alto Huallaga (30) (Table 1). We isolated 211 Gram-negative bacterial isolates from the surface-sterilized roots; however, only 78 strains (36% of the total) showed nitrogen-fixing ability in our tests. As we were focusing on B. vietnamiensis, a nitrogen-fixing species, only they were kept isolated for further analysis. Most (44, 57.90%) of these Gram-negative and diazotrophic isolates came from plants grown in the Huallaga Central Valley, while only 6 came from plants grown in the Alto Mayo Valley.
| Valley | Province | District/Sector | Rice Cultivar | Geographical Coordinates | Amount of Samples | Number and code of Diazotrophic Gram-negative Strains |
|---|---|---|---|---|---|---|
| Bajo Mayo | Lamas | Cacatachi/Rosanaico | La Conquista | 6° 28' 11” S, 76° 26' 26” W; 299 m.a.s.l. | 30 | (3)-B1; B3; B13 |
| San Martín | Juan Guerra/Estación El Porvenir | La Conquista, Valor, La Esperanza, Fedearroz 60 | 6° 35' 49” S, 76° 19' 32” W; 309 m.a.s.l. | (4)-B34; B37; B48; B49 | ||
| Alto Mayo | Rioja | Awajun/San Francisco de Amayo | Valor, La Esperanza, La Victoria | 5° 46' 43” S, 77° 18' 29” W; 916 m.a.s.l. | 36 | (3)-B51; B52; B74 |
| Moyobamba | Moyobamba/La conquista | Valor | 5° 52' 41” S, 77° 10' 10” W; 913 m.a.s.l. | 21 | (3)-B61; B89; B90 | |
| Huallaga Central | Bellavista | Bellavista/El Porvenir | Ferón | 7° 03' 11” S, 76° 33' 54” W; 235 m.a.s.l. | 15 | (9)-B91; B92; B94; B102; B103; B104; B105; B106; B106a |
| Bellavista | San Rafael/Carhuapoma | La Esperanza | 7° 00' 21” S, 76° 30' 08” W; 228 m.a.s.l. | (9)-B126; B129; B130; B131; B135; B138; B139; B140; B142 | ||
| Picota | Picota/Santa Rocillo | Valor | 6° 55' 32” S, 76° 22' 23” W; 218 m.a.s.l. | (21)-B107; B108; B109; B110; B111; B113; B114; B115; B118; B119; B121; B122; B124; B154; B155a; B158; B159; B160; B161; B162; B163 | ||
| Picota | San Hilarión/San Hilarión | Ferón | 6° 59' 55” S, 76° 27' 05” W; 225 m.a.s.l. | (5)-B145; B146; B147; B151; B152 | ||
| Alto Huallaga | Tocache | Uchiza/Nueva Esperanza | El Valor | 8° 15' 39” S, 76° 33' 59” W, 495 m.a.s.l. | 15 | (7)-B167; B168a; B168b; B169a; B169b; B171a; B172a |
| Uchiza/San Juan de Porongo | El Valor, Ferón | 8° 23' 27” S, 76° 17' 17” W, 625 m.a.s.l. | (10)-B172b; B175; B176; B177; B178; B188; B189; B190; B191; B192 | |||
| Tocache/San Miguel del Porvenir | Ferón, Valor | 8° 16' 26” S, 76° 32' 25” W; 590 m.a.s.l. | 15 | (4)-B193; B195; B196; B201 |
3.2. Molecular Discrimination of Burkholderia
Only 4 isolates (5.13%) from our collection of Gram-negative and nitrogen-fixing bacteria, together with two previously isolated strains of B. vietnamiensis la3c3 and la1a4 [34], amplified a fragment of 704 bp corresponding to the recA gene using primers specific to Burkholderia. Sequencing of the recA gene confirmed the taxonomic identity of the isolates in the species B. vietnamiensis. These 4 isolates all originated from the Alto Huallaga Valley and, more specifically, from the Nueva Esperanza sector of the Uchiza district. The la3c3 and la1a4 strains were originally sampled from the Central Huallaga Valley [34].
3.3. BOX-PCR Genomic Profiling
To study the intraspecific diversity of the six B. vietnamiensis strains, genomic DNA profiles were performed using BOX-PCR analysis. The fragments obtained ranged from 500 to 10,000 bp. Using a similarity level of 90% as a cut-off point, 5 different groups were obtained (Fig. 1). These groups were named from G1 (BOX group 1) to G5 (BOX group 5). Each group, except G1, contains only one isolate. The strains la3c3 and la1a4 fall into two different BOX-PCR groups. Based on the BOX-PCR dendrogram, one strain per group was selected for amplification, sequencing, and phylogenetic study of the recA gene.
3.4. Phylogeny of the recA Gene
Sequencing resulted in partial recA sequences of 625-667 bp. Based on blast and phylogenetic analyses, it was confirmed that the 5 isolates representing the BOX-PCR groups belonged to the species B. vietnamiensis, with percentages of similarity of the recA gene between 98.36 and 99.51% with the type strain of B. vietnamiensis LMG 10929T. The maximum likelihood (ML) phylogenetic tree, including sequences of the type strain of the genus Burkholderia sensu stricto, is shown in Fig. (2). All B. vietnamiensis sequences were clustered in the same highly supported (99%BP) clade. The recA gene sequences have been deposited in GenBank under the following accession numbers: OQ706314-OQ706318.
3.5. Growth Promotion Mechanisms Detected in Burkholderia vietnamiensis
All the strains studied were able to express the direct mechanisms of growth promotion, auxin, and siderophore production, phosphate solubilisation, and diazotrophic capacity (Table 2), as well as the capacity for antibiosis and antagonism against phytopathogenic rice fungi as indirect growth promotion mechanisms (Table 3). All strains were able to produce auxin in the basal TSB medium (0 µg mL-1) and the TSB media supplemented with tryptophan (100 ppm to 600 ppm). There was also a strong positive correlation between auxin production and tryptophan supplementation in TSB broth (r2= 0.99). Under all conditions, B. vietnamiensis strain B169b produced a significantly higher amount of auxin compared to the other strains.
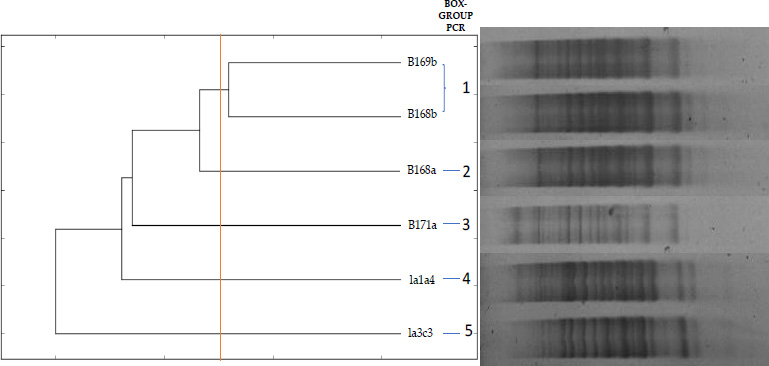
Fingerprinting dendrogram of similarity based on the BOX-PCR profiles of the isolates of this study using the UPGMA algorithm and the Jaccard coefficient with 2% tolerance using the online dendrogram construction utility DendroUPGMA (http://genomes.urv.es/UPGMA/) [47].
All strains were efficient in the production of siderophores, forming orange halos around the bacterial colonies of smaller or larger diameter depending on the strain (Fig. 3), highlighting B. vietnamiensis strain B169b as significantly the most efficient strain (139.52%). Regarding phosphate solubilization, all strains were able to solubilize tricalcium phosphate from NBRIP agar, as evidenced by the appearance of a transparent halo around the bacterial colonies.
This activity was reflected in the solubilization indices and the solubilization efficiency obtained, although no statistically significant differences were observed between them. The strains also showed no statistically significant differences in the concentration of phosphate released into the culture medium. A negative correlation was observed between the final pH of the culture medium and the difference in soluble phosphate in the culture medium with the white treatment (r2= -0.92). Finally, all strains were able to form a sub film in the semi-solid JMV medium and to grow on the JMV agar, both nitrogen-free, indirectly demonstrating their ability to fix atmospheric nitrogen.
3.6. Antagonism and Antibiosis Capacity of B. vietnamiensis Strains
In terms of antibiosis capacity, all strains inhibited the growth of the five phytopathogenic fungi, but strain la3c3 showed the greatest antibiosis activity, inhibiting 100% of the growth of R. solani, R. oryzae, N. oryzae and N. sigmoidea. All five strains were able to inhibit 100% of the growth of N. oryzae, the most susceptible phytopathogenic fungus, while the least susceptible was Rhizoctonia oryzae-sativae, with radial growth inhibition ranging from 76.47% (la1a4) to 86.04% (B171a) (Table 3 and Fig. 4).
All strains inhibited the growth of the five phytopathogenic fungi, with strain B169b showing the highest range of antagonistic capacity against the five phytopathogenic fungi. The most susceptible fungus was N. sigmoidea, with a range of 59.94% (la3c3) to 77.56% (B169b) antagonism, and the least susceptible was N. oryzae, with a range of 41.98% (B168a) to 49.21% (la1a4) growth inhibition (Table 3 and Fig. 3).
All B. vietnamiensis strains increased length, germination, and vigor index in both rice cultivars under gnotobiotic conditions, although there was no clear significant difference among isolates. While there were substantial differences between the strains in terms of root length in the LE cultivar, we found none for LC. The same result was observed for the length of aerial parts. The highest growth promotion effects compared to the uninoculated reached 50% and 35.6% for root length (LE/B169b and LC/lac3c, respectively) and 32.2% and 33.4% for shoot length (LE/B168a and LC/B168a, respectively).
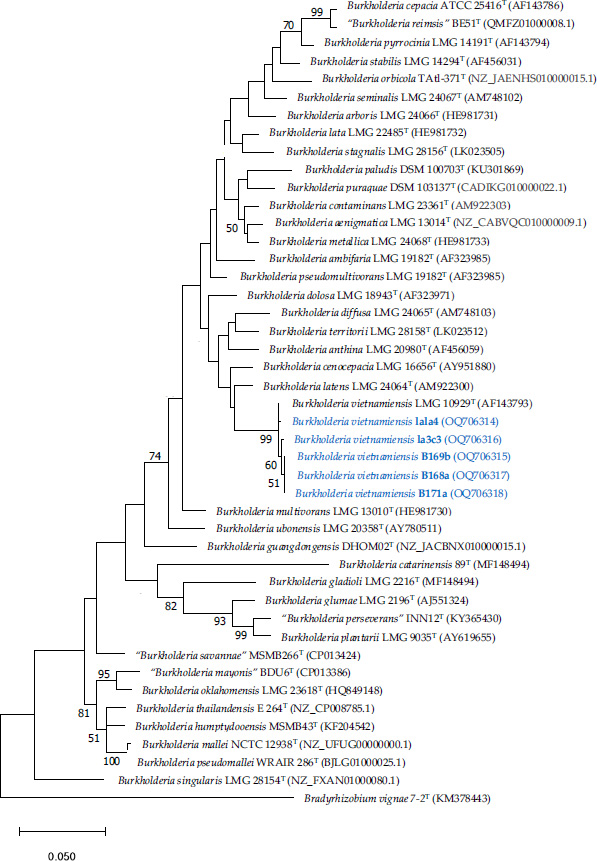
Maximum-likelihood phylogenetic tree based on recA gene sequences (650 positions) showing the relationships among phytobeneficial Burkholderia isolated in this study and closely related species of the genus Burkholderia sensu stricto. The significance of each branch is indicated by a bootstrap value (as a percentage) calculated for 1000 subsets (only values greater than 50% are indicated). Bar, 5 substitutions per 100 nucleotide positions. The recA sequence of Bradyrhizobium vignae 7-2T was used as an outgroup.
| Strains | Auxins Production (µg mL-1) | Siderophores Production | Phosphate Solubilization | Free-living Nitrogen Fixation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 ppm | 100 ppm | 200 ppm | 400 ppm | 600 ppm | EPS (%) | Qualitative | Quantitative | ||||
| Solubilization Index | Solubilization Efficiency | pH | Solubilized P (mg PO4 mL-1) | ||||||||
| la1a4 | 8.10 (±0.15) B | 8.86 (±0.21) B | 8.72 (±0.16) B | 11.79 (±0.35) A | 10.97 (±0.20) B | 93.38 (±1.85) B | 2.63 (±0.04) A | 162.73 (±3.54) A | 3.89 | 11.62 (±0.22) A | + |
| la3c3 | 0.25 (±0.25) D | 0.70 (±0.03) D | 0.15 (±0.08) D | 1.27 (±0.10) C | 2.76 (±0.05) D | 52.32 (±1.18) D | 2.64 (±0.03) A | 163.88 (±3.46) A | 3.93 | 12.18 (±0.77) A | + |
| B169b | 9.08 (±0.07) A | 9.70 (±0.17) A | 13.98 (±0.31) A | 11.76 (±0.07) A | 13.90 (±0.22) A | 139.52 (±3.30) A | 2.58 (±0.04) A | 157.80 (±4.33) A | 3.81 | 15.99 (±0.58) A | + |
| B168a | 0.10 (±0.05) D | 1.89 (±0.09) C | 1.41 (±0.05) C | 3.79 (±0.10) B | 4.60 (±0.44) C | 72.3 (±11.91) C | 2.59 (±0.04) A | 159.31 (±3.66) A | 3.84 | 15.26 (±0.28) A | + |
| B171a | 2.82 (±0.05) C | 1.41 (±0.05) C | 0.62 (±0.03) D | 3.30 (±0.07) B | 3.33 (±0.38) B | 47.72 (±1.87) DE | 2.53 (±0.01) A | 153.25 (±0.69) A | 3.69 | 17.63 (±0.48) A | + |
| C.V (%) | 5.94 | 5.01 | 5.68 | 4.76 | 7.09 | 10.03 | 2.65 | 4.24 | -- | 19.30 | -- |
| Strains | Rhizoctonia solani (RS1) | Rhizoctonia oryzae (RO1) | Rhizoctonia oryzae-sativae (ROS1) | Nigrospora oryzae (NO1) | Nakataea sigmoidea (NS1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANTIBIOTIC (%) | |||||||||||||||||
| B168a | 78.59 (±0.12) C | 100.00 (±0.00) A | 82.23 (±1.94) AB | 100.00 (±0.00) A | 100.00 (±0.00) A | ||||||||||||
| B171a | 100.00 (±0.00) A | 100.00 (±0.00) A | 86.04 (±0.35) A | 100.00 (±0.00) A | 90.86 (±0.24) B | ||||||||||||
| B169b | 100.00 (±0.00) A | 77.93 (±2.04) B | 79.72 (±1.53) BC | 100.00 (±0.00) A | 87.80 (±0.50) C | ||||||||||||
| la3c3 | 100.00 (±0.00) A | 100.00 (±0.00) A | 84.82 (±0.69) A | 100.00 (±0.00) A | 100.00 (±0.00) A | ||||||||||||
| la1a4 | 80.39 (±0.61) B | 75.80 (±3.49) B | 76.47 (±0.80) C | 100.00 (±0.00) A | 100.00 (±0.00) A | ||||||||||||
| C.V(%) | 0.39 | 3.45 | 2.56 | 0 | 0.45 | ||||||||||||
| ANTAGONISM (%) | |||||||||||||||||
| B168a | 56.22 (±2.60) A | 59.03 (±2.36) A | 73.85 (±5.87) A | 41.98 (±1.92) A | 73.63 (±1.46) A | ||||||||||||
| B171a | 61.20 (±2.64) A | 57.54 (±1.47) A | 54.39 (±5.27) B | 44.96 (±1.20) A | 72.79 (±2.00) A | ||||||||||||
| B169b | 58.71 (±1.00) A | 55.06 (±2.27) A | 65.87 (±2.89) AB | 47.50 (±3.07) A | 77.56 (±1.45) A | ||||||||||||
| la3c3 | 64.68 (±2.13) A | 61.51 (±0.99) A | 64.87 (±2.00) AB | 44.72 (±1.61) A | 59.94 (±0.92) B | ||||||||||||
| la1a4 | 56.22 (±1.42) A | 57.54 (±0.99) A | 61.87 (±2.40) AB | 49.21 (±3.17) A | 63.75 (±2.73) B | ||||||||||||
| C.V(%) | 8.52 | 7.26 | 15.3 | 12.5 | 6.4 | ||||||||||||
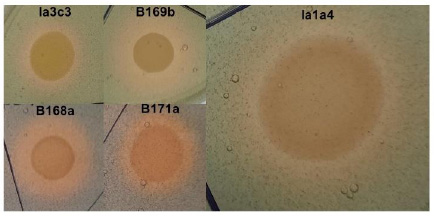
Production of siderophores by Burkholderia vietnamiensis strains on CAS agar
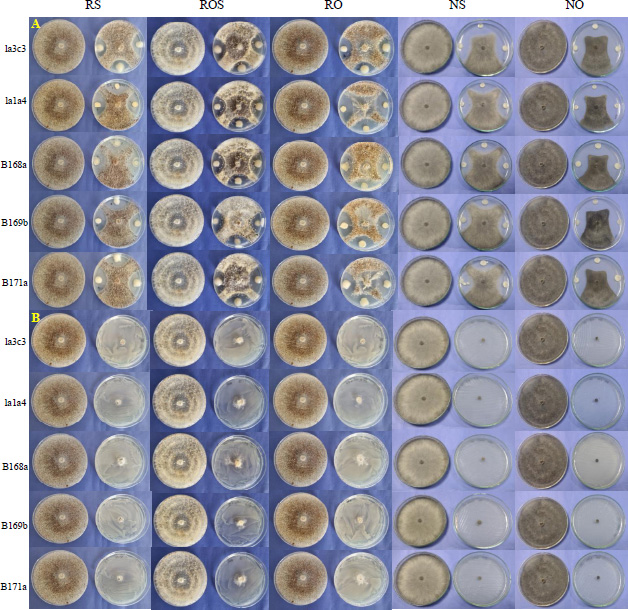
Evaluation of antagonism (A) and antibiosis (B) in five strains of Burkholderia vietnamiensis against five strains of phytopathogenic rice fungi, RS1 (Rhizoctonia solani), ROS1 (Rhizoctonia oryzae-sativae), RO1 (Rhizoctonia oryzae), NS1 (Nakataea sigmoidea) and NO1 (Nigrospora oryzae).
All inoculation conditions resulted in a significant increase in the total length or vigor index of the plant, irrespective of the cultivar, but with no significant difference between the cultivars (Table 4). While the germination rate differed between the two varieties (p<0.04), the effects of inoculation on this rate were highly significant compared to the uninoculated conditions (p<0.003), with a higher germination rate for inoculated seeds.
Finally, in the 21-day test, there was no significant difference between the strains of B. vietnamiensis with P. megaterium SMBH14-02 for length, dry weight, and nitrogen accumulation. Similarly, non-significant differences were observed in the accumulation of TDW inoculated with B. vietnamiensis strains in the LC cultivar. Differences were observed in the accumulation of total nitrogen, which was higher in the LE cultivar inoculated with B. vietnamiensis la3c3 (6.96%) and with B. vietnamiensis B171a (7.17%) in the LC cultivar (Table 5).
| Strains | RL | SL | TL | Germination | Vigor Index | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| LE1 | LC1 | LE1 | LC1 | LE1 | LC1 | LE | LC | LE1 | LC1 | |
| la1a4 | 58.63 (±1.42) DE | 60.41 (±0.92) A | 38.16 (±1.42) C | 45.36 (±1.09) A | 94.40 (±2.57) B | 105.77 (±1.71) A | 98 | 98 | 8582.06 (±199.35) Bb | 10365.66 (±194.20) Aa |
| la3c3 | 68.13 (±1.02) C | 60.52 (±2.19) A | 44.97 (±0.94) AB | 44.73 (±2.25) A | 112.71 (±2.02) A | 105.25 (±3.54) A | 100 | 94 | 11310.27 (±193.25) Aa | 9892.84 (±414.10) Ab |
| B169b | 75.27 (±1.97) A | 58.74 (±1.88) A | 44.22 (±1.23) B | 44.20 (±1.46) A | 115.18 (±2.79) A | 102.94 (±3.18) A | 96 | 94 | 11470.50 (±305.63) Aa | 9675.80 (±308.90) Ab |
| B168a | 65.22 (±1.62) CD | 58.79 (±1.52) A | 49.75 (±1.33) A | 45.62 (±2.20) A | 111.12 (±2.01) A | 104.41 (±4.09) A | 100 | 96 | 11496.67 (±294.38) Aa | 10023.49 (±355.84) Ab |
| B171a | 72.8 ((±1.80) B | 57.89 (±1.43) A | 43.47 (±0.96) B | 42.65 (±1.40) A | 112.08 (±2.26) A | 100.54 (±2.49) A | 100 | 94 | 11626.93 (±272.51) Aa | 9450.57 (±264.82) Ab |
| Priestia megaterium SMBH14-02 | 67.81 (±0.82) CD | 60.68 (±1.31) A | 42.28 (±0.91) BC | 40.36 (±1.10) AB | 108.21 (±1.60) A | 101.04 (±2.16) A | 100 | 100 | 11008.73 (±172.29) Aa | 10103.97 (±236.04) Ab |
| Non-inoculated control | 49.97 (±1.11) E | 44.63 (±1.19) B | 37.61 (±0.70) DE | 34.19 (±1.22) B | 88.32 (±1.59) B | 78.82 (±2.01) B | 92 | 84 | 8235.35 (±176.77) Ba | 6620.43 (±201.69) Bb |
| CV (%) | 12.13 | 14.73 | 14.83 | 20.63 | 11.17 | 15.76 | 12.31 | 16.94 | ||
| Values followed by different uppercase letters indicate significant differences between strains, and values followed by different lowercase superscript letters indicate significant differences between cultivars. RL: Root length; SL: Shoot length; TL: Total length of the plant; LE: INIA 507 “La Esperanza” rice cultivar; LC: INIA 510 “La Conquista” rice cultivar 1Kruskal Wallis test, p = 0.05. |
||||||||||
| Parameter | Burkholderia vietnamiensis | Priestia Megaterium SMBH14-02 | Non-inoculated Control | CV (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| la1a4 | la3c3 | B169b | B168a | B171a | |||||
| RL-LE2 | mm. | 103.89 (±3.22) B | 103.90 (±5.02) B | 115.78 (±5.15) B | 142.04 (±1.90) A | 116.04 (±2.98) B | 106.10 (±7.64) B | 97.51 (±2.98) B | 13.28 |
| RL-LC2 | 73.94 (±4.94) AB | 77.36 (±4.83) AB | 79.68 (±3.04) A | 82.93 (±3.00) A | 88.55 (±2.14) A | 81.77 (±1.64) A | 63.86 (±3.10) B | 13.90 | |
| SL-LE2 | 167.64 (±2.59) B | 192.27 (±3.76) A | 175.77 (±4.91) AB | 121.85 (±2.72) C | 169.79 (±4.50) B | 172.00 (±7.61) B | 159.81 (±2.68) B | 8.47 | |
| SL-LC1 | 196.73 (±5.44) AB | 201.12 (±7.77) AB | 195.07 (±3.84) AB | 211.53 (±4.89) A | 199.70 (±3.27) AB | 205.49 (±5.23) AB | 180.50 (±9.57) B | 9.67 | |
| TL-LE1 | 268.29 (±3.42) ABCa | 285.26 (±5.30) Aba | 288.06 (±8.96) Aa | 258.98 (±4.28) BCb | 281.65 (±5.50) Aa | 263.82 (±8.74) ABCb | 251.13 (±4.81) Ca | 7.22 | |
| TL-LC2 | 275.69 (±8.02) Aa | 281.12 (±10.43) Aa | 265.25 (±5.17) Ab | 289.96 (±8.96) Aa | 284.90 (±4.73) Aa | 290.04 (±4.63) Aa | 232.43 (±13.65) Ba | 9.41 | |
| RDW-LE2 | mg | 11.27 (±0.18) A | 11.35 (±0.29) A | 11.28 (±0.40) AB | 11.25 (±6.05) A | 11.57 (±0.20) A | 9.81 (±0.27) B | 9.31 (±0.18) B | 7.84 |
| RDW-LC1 | 10.58 (±0.32) A | 9.98 (±0.30) ABC | 10.01 (±0.27) ABC | 10.56 (±0.19) AB | 11.21 (±0.26) A | 9.69 (±0.47) BC | 8.77 (±0.20) C | 9.36 | |
| SDW-LE2 | 16.85 (±0.61) AB | 17.55 (±0.34) AB | 17.69 (±0.60) AB | 16.47 (±0.81) B | 19.20 (±0.64) A | 13.37 (±0.48) C | 13.39 (±0.35) C | 11.01 | |
| SDW-LC1 | 14.79 (±0.54) AB | 13.95 (±0.91) ABC | 14.36 (±0.41) ABC | 13.78 (±0.34) ABC | 16.47 (±0.75) A | 12.60 (±0.49) BC | 11.68 (±0.86) C | 14.76 | |
| TDW-LE2 | 27.85 (±0.68) Aa | 28.61 (±0.50) Aa | 28.78 (±1.04) Aa | 27.47 (±0.90) Aa | 30.49 (±2.54) Aa | 22.47 (±0.75) Ba | 23.05 (±0.49) Ba | 8.91 | |
| TDW-LC1 | 24.62 (±0.43) ABb | 22.16 (±0.61) BCb | 23.44 (±0.43) ABa | 24.72 (±0.72) ABb | 26.20 (±1.01) Ab | 22.73 (±0.62) BCa | 20.44 (±0.59) Cb | 8.84 | |
| % NPA-LE | % | 4.91 | 5.67 | 4.21 | 4.64 | 4.76 | 4.56 | 4.29 | --- |
| %NPA-LC | 4.63 | 4.87 | 5.03 | 4.06 | 6.24 | 4.39 | 4.18 | --- | |
| % NPR-LE | 1.80 | 1.29 | 1.39 | 1.31 | 1.76 | 1.34 | 1.05 | --- | |
| % NPR-LC | 1.54 | 1.31 | 1.44 | 1.78 | 0.93 | 1.25 | 1.54 | --- | |
| %NT-LE | 6.70 | 6.96 | 5.60 | 5.95 | 6.52 | 5.90 | 5.34 | --- | |
| %NT-LC | 6.17 | 6.19 | 6.46 | 5.83 | 7.17 | 5.64 | 5.71 | --- | |
4. DISCUSSION
Root-associated bacteria are an important part of the phytomicrobiome, not only because they represent a selected fraction of the soil microbiota but also because of the growth-promoting mechanisms they express in close association with plants, especially in the Poaceae family. There are several reports of bacteria being selected for their diazotrophic capacity in rice cultivation [36, 38, 68-71].
De Oliveira et al. [68], in a bioprospecting study on diazotrophic bacteria in the rice cultivar BRS tropical, reported that 18% of the isolates obtained were similar to those in the genus Burkholderia, and diverse species of Burkholderia have been repeatedly isolated from healthy rice roots [66]. In our study, the proportion of Burkholderia was much lower (5.26%) and was isolated only from the rice variety “El valor”, grown in the Alto Huallaga valley. It is worth mentioning that the primers used for the detection of Burkholderia missed some isolates and did not amplify them. However, when searching the complete or draft genomes of B. vietnamiensis available in NCBI, we did not find any example of genomes (among the 208 genomes available) where the primers would not have amplified the recA fragment.
This explanation, therefore, seems unlikely. The low frequency of Burkholderia isolates in our study could also be due to the selection of isolates for which we attempted to amplify a recA fragment. We focused on isolates that could fix nitrogen because all previous studies and available genomes have shown that B. vietnamiensis strains are free-living nitrogen fixers. This choice may have excluded some other Burkholderia species that are not free-living nitrogen fixers, thus reducing the number of Burkholderia isolates. However, several have been shown to act as plant-associated nitrogen fixers [72]. Finally, the low frequency of B. vietnamiensis isolates in our sampling may simply reflect their low frequency in soil. However, this species has been shown to be frequently recovered from rice roots in different geographical locations. Moreover, Burkholderia s. l. is a highly conserved microbial group of the microbiota in rice, regardless of soil type and cultivar [73].
Quite surprisingly, the box PCR profiles showed diversity among the different isolates and the four recovered in this study. It is expected that a very small number of isolates recovered from a single site would end up as a single identical clone, which is not the case since we detected at least three different profiles (together with the two other strains isolated in a previous study, but from a different site in Peru). The recA gene, as a reliable phylogenetic marker for assigning taxonomic identity within the genus Burkholderia [10, 74-78], confirmed that the 4 strains isolated belonged to the species B. vietnamiensis but showed less diversity than the Box-PCR approach (as expected). In fact, the 3 representative isolates of the 3 groups showed the same recA sequences, while the other 2 showed 2 nucleotide differences with our isolates. The Box-PCR diversity, with 3 different profiles among the 4 isolates obtained, however, suggests that we may indeed have missed some B. vietnamiensis diversity in our sampling and that we should possibly intensify our efforts to recover more isolates.
Based on these results and the genetic differences detected between the isolates, we expected some discrepancies in their growth-promoting abilities.
While several studies have previously demonstrated the beneficial effects of B. vietnamiensis on rice growth and possibly protection against pathogens, we investigated whether and how these strains might actually play a role as growth promoters. Among bacterial auxins, indoleacetic acid (IAA) is one of the most studied, influencing root architecture, nutrient uptake, and tolerance to abiotic factors. Bacterial IAA concentrations are influenced by intrinsic factors, such as strain type, presence of complete biosynthetic pathway genes, or extrinsic factors, such as precursor concentration, pH, and carbon source, among others [79]. It has been reported that IAA production is common in plant-associated strains of B. vietnamiensis [24, 30, 33, 34, 68, 80, 81]. Our results showed that there is a wide range of variation in auxin production among our isolates. At 600 ppm L-tryptophan, strain B169b produced 5 times more auxin than strain la3c3 (13.90 vs 2.76 µg mL-1). Most of the auxin found in the rhizosphere is believed to come from biosynthesis by microorganisms [82]. Variations in auxin production between strains present in the rhizosphere can, therefore, have a major impact on plant development, and from the point of view of artificial inoculation, our results proved that there is a basis for bacterial selection of the most productive strains.
B. vietnamiensis is also characterized by the production of siderophores [25, 81, 83, 84]. Among the functions performed by siderophores, biocontrol stands out due to their high affinity for Fe3+, which limits their access to phytopathogens and promotes plant growth under iron-limiting conditions [19]. As for auxin production, we also found large differences in the production of siderophores among the isolates, ranging from 47.72% (B171a) to 139.52% (B169b), leaving for auxin the possibility of strain selection for their ability to produce the highest amount of siderophore that can play a major role in agriculture [85].
Regarding the other two activities, we did not detect significant differences in phosphate solubilization, although all strains showed qualitative and quantitative evidence of P solubilization. The ability of B. vietnamiensis to solubilize insoluble phosphates has been widely reported previously [33, 36, 37, 81, 86]. Estrada et al. [33] reported that B. vietnamiensis isolates from rice achieved solubilisation index values similar to those reported in this study (2.34-2.95). However, while Bashan et al. [87] suggested a low correlation between halo formation and the ability to solubilise phosphates released in the liquid medium, we observed in our study that all strains that formed a solubilization halo (qualitatively) also solubilized phosphate in liquid medium (quantitatively).
Finally, all isolates showed the ability to fix nitrogen as free-living bacteria, but we could not measure this activity precisely and, therefore, could not estimate significant differences between them.
Given these results, we expected not only an effect of inoculation on plant growth but also possibly differences between strains for plant phenotypic traits in response at least to auxin production level differences. The beneficial interaction between B. vietnamiensis and rice has been described previously [12, 24, 25, 31, 34, 88], but with sometimes conflicting results. King et al. [89] reported that Nipponbare rice (japonica) plants inoculated with B. vietnamiensis TVV75 (LMG10929T) had no significant effect on biomass production compared to the non-inoculated control at 7 and 14 days after inoculation under gnotobiotic conditions. In our study, we found significant differences between the strains and the non-inoculated control evaluated at 10 and 21 days after inoculation. The LE genotype showed significant differences for root and shoot length at 10 days between isolates but with a weak correlation between the two sets of measurements (r2=0.38), while the LC genotype showed almost no significant differences, but with a higher r2 value (0.75). The low correlation value for LE resulted in a lack of significant differences between the strains when considering the total length (shoot + aerial), except for isolate la1a4, which is not different from the uninoculated condition. However, it was one of the higher auxins and siderophore producers in our tests. Many previous studies demonstrated the effect of bacterial auxin on plant growth in terms of height, root length, dry weight shoot and dry weight root, plant nutrient content, chlorophyll content, leaf area, and yield [90], with an effect being concentration-dependent [91]. We did not detect any relation between the level of auxin production of each strain and their impact on either root or shoot length. There was either no significant difference among strains (for variety LC) or no correlation for variety LE. The other putative growth-promoting activities we measured in our isolates do not appear to have an effect on rice growth either. This lack of correlation may be due to the absence of a dose effect, which would be rather surprising given the different production levels. More likely, the growing conditions may not have allowed auxin-related effects to be fully expressed. In our study, we chose to place our plants in non-limiting growing conditions, i.e., with a rich growth medium, where Fe or phosphate is not limiting and, therefore, the capacity of the bacteria to recover these elements does not play a major role. The increase in plant height and biomass is associated with an increase in the expression of plant genes involved in iron storage, siderophore biosynthesis, and nutrient transport [92].
One exception is related to nitrogen. It has been reported that inoculation of rice with B. vietnamiensis increases the efficient use of mineral nitrogen [24, 34]. Consequently, the five treatments with B. vietnamiensis and the control strain P. megaterium SMBH14-02, inoculated in the two rice varieties and supplemented with mineral nitrogen (KNO3 1%), increased the nitrogen content in the aerial and root parts from 11 to 19% on average. Although nitrogen fixation is an intrinsic capacity of B. vietnamiensis [25, 29, 93], recent studies have concluded that it would not be the main growth-promoting mechanism in rice. Shinjo et al. [24] inoculated B. vietnamiensis RS1 into seedlings of rice cultivar Nipponbare (japonica) and concluded that the promotion of root growth occurred through hormonal regulation and increased nitrogen uptake through the overexpression of genes related to the uptake, transport, and assimilation of mineral nitrogen. This ability is intrinsic to root-associated Burkholderia, as it improves the acquisition and transport of nutrients in tissues at the transcriptional level [92].
Finally, with regard to plant genotype, it is well known that plant genotype influences the response to inoculation of specific bacterial strains [94-96]. Wallner et al. [12] inoculated B. vietnamiensis LMG10929 into two rice cultivars, Nipponbare (japonica) and IR64 (indica); although they observed similar colonization patterns between the two genotypes, they found that there was a strong influence of rice genotype, particularly in relation to the indica group more than the japonica group. The IR64 (indica) cultivar inoculated with B. vietnamiensis increased the expression of the nitrate transporter NRT1.1B, affecting its capacity for assimilation and uptake of nitrate from the nutrient solution. In our study, the results suggested, once again, as in other studies, that selecting bacterial strains for inoculation to improve plant growth can only be done by taking into account the diversity of cultivated varieties and also by integrating into breeding programs the ability of varieties to interact with micro-organisms.
4.1. Inoculation of B. vietnamiensis and Protection against Pathogens
Recently, Wang et al. [97] reported that the inoculation of B. vietnamiensis strain B23 in Citrus plants increased the expression of genes related to the use and uptake of nutrients from the rhizosphere and increased antagonistic activities against competing bacteria and fungi, as well as resistance to competitor-derived metabolites. Meng et al. [28] reported the isolation of B. vietnamiensis C12 as an antifungal bacteria of the medicinal plant Ficus tikoua and demonstrated the production of siderophores with bactericidal activity, ornibactin C-4 and C-8. Similarly, Wang et al. [98] demonstrated the ability of B. vietnamiensis YQ9 to produce hydroxamate type siderophores.
All B. vietnamiensis strains in our study showed antagonistic and antibacterial activity against three members of the Rhizoctonia complex, including Rhizoctonia solani, the causal agent of rice downy mildew [28, 34, 99-101]. The antifungal activity of B. vietnamiensis against Nakataea sigmoidea and Nigrospora oryzae, the etiological agents of stem rot [103] and panicle branch rot [102], respectively, is reported for the first time. Meng et al. [28] reported the antifungal activity of B. vietnamiensis C12 against several phytopathogenic fungi, reaching 94.78% inhibition against R. solani, a higher range than that reported in our study (56.22 to 64.68%). The authors concluded that this antifungal activity against R. solani is due to the production of several secondary metabolites, such as siderophores ornibactin C4 and C8, the antifungal peptides burkholdin 1097, 1213, 1215, and 1119, and the monoterpenoid phenol carvacrol. In this study, it was found that all B. vietnamiensis strains have a strong effect on reducing fungal growth on artificial media in Petri dishes. Such ability of Burkholderia isolates to directly suppress the growth of phytopathogenic fungi has been described previously [90]. Antifungal metabolites produced by Burkholderia that are effective against the Rhizoctonia complex include pyrrolnitrin, phenazines, 1-phenazine carboxylic acid, volatile indolic compounds [104], siderophores, such as ornibactin [105], some quinolone antibiotics, such as hydroxymethyl-alkylquinoline [106], and occidiofungins A-D [107]. Searches for these compounds in B. vietnamiensis genomes have detected some of the genes involved in the production of these compounds (such as ornibactin), while others were not detected (such as pyrrolnitrin or occidiofungins A-D).
CONCLUSION
Burkholderia vietnamiensis, associated with rice grown in the Peruvian highland forest, presents a great genetic diversity and expresses different mechanisms of direct and indirect plant growth promotion. It exhibits great potential in the production of auxins, siderophores, phosphate solubilization, and the promotion of vigor index in rice. There is still a long way to isolate and characterize the antifungal compounds produced by B. vietnamiensis, but we can expect new metabolites to be found. It is also clear that there is a huge gap between Petri dish tests and the use of either live bacteria or metabolite extract in the field to combat rice diseases. However, even if Burkholderia s.s. is still a problematic genus for use as a bioinoculant, and B. vietnamiensis species in particular, it remains an extremely interesting and promising genus in the search for solutions and alternative compounds to fight against rice diseases
AUTHORS’ CONTRIBUTION
It is hereby acknowledged that all authors have accepted responsibility for the manuscript's content and consented to its submission. They have meticulously reviewed all results and unanimously approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| s. l | = sensu lato |
| s. s | = sensu stricto |
| OD | = Optic density |
| LE | = La Esperanza |
| LC | = La Conquista |
| TDW | = Total Dry Weight |
| PCR | = Polymerase Chain Reaction |
| IAA | = Indole Acetic Acid |
AVAILABILITY OF DATA AND MATERIALS
All data and information supporting the research are provided within the article. In particular the sequence data for the recA gene (OQ706314 OQ706318) are available at https://www.ncbi.nlm.nih.gov/.
FUNDING
The study was funded by the Fondo National de Desarrollo Científico, Tecnológico y de Innovación Tecnológica (FONDECyT) (grant number CONTRATO N° 062-2021-FONDECyT).
ACKNOWLEDGEMENTS
The authors would like to thank the rice specialist Edson Torres Chavez for donating seeds of the La Esperanza and La Conquista varieties for the promotion of inoculation tests. They would also like to thank Dr. Miguel Ángel Garrido Rondoy, Head of the Plant Disease Diagnostic Center, Faculty of Agricultural Sciences, National University of Tumbes, who donated the strains of phytopathogenic fungi used in the antagonism and antibiosis tests. We thank the National University of Barranca for the funding provided for the publication of this manuscript.


