All published articles of this journal are available on ScienceDirect.
Biotechnological Approaches to Increase the Bacterial and Fungal Disease Resistance in Potato
Abstract
Potato (Solanum tuberosum) is a valuable cultivated vegetable with high nutritional qualities containing a variety of essential ingredients such as starch, vitamins and essential minerals. However cultivating potatoes is significantly complicated by diseases caused by a number of phytopathogens of both bacterial and fungal nature. Phytophthora infestans, Alternaria sp., Phoma exigua, Rhizoctonia solani, Fusarium sp., Verticillium sp., Clavibacter michiganensis subsp. sepedonicus, Pectobacterium sp., Dickeya sp. are considered amongst the most dangerous phytopathogens.
This paper reviews the latest biotechnological approaches to the creation of potato plants resistant to a wide range of pathogens. In particular, the possibilities of applying genetic engineering methods to obtain potato plants resistant to pathogens, such as Solanum venture, S. mochiquence, S. demissum, S. bulbocastanum; avirulence genes; genes of antimicrobial peptides of plant, bacterial and animal origin by transferring the genes of resistance (R-genes) isolated from systematically related species into their genome. The review states that marker-assisted selection is suitable for obtaining varieties of S. tuberosum resistant to bacterial and fungal phytopathogens, where the R-genes or QTL regions can act as markers. Prospects for the use of genome editing technology using CRISPR/Cas9 or TALEN systems as one of the newest approaches to creating phytopathogen-resistant S. tuberosum plants have been considered. Achievements and successes in this way using these methods are analyzed in a detailed way in this review.
1. INTRODUCTION
Potatoes (Solanum tuberosum L.) occupy one of the leading positions in the world food market among vegetable crops [1-3]. These plants are a valuable food crop because they are consumed unprocessed (table varieties), serve as raw materials for manufacturing semi-finished products (french fries), dehydrated products (potato flour, potato flakes) and snacks (potato chips, potato crisps) [4]. Potatoes are also used to make starch, glucose, dextrin, etc. Besides the food industry, potato starch is also used in the production of paper, fabrics, alcoholic beverages, etc. Potato processing waste is a useful fodder for livestock [4].
S. tuberosum is characterized by a high content of nutrients, trace elements and biologically active compounds. Depending on the crop variety, the starch content can range from 13 to more than 30%. Also, potatoes are an important source of vitamins and minerals, including potassium, magnesium, iron. Among vitamins, the highest content is of vitamin C (27 mg per 100 g of tubers) and vitamins of group B (B1, B2, B6, B9). About 300 g of potatoes contain the daily requirement of vitamin C for humans, which is very important during the winter season when the diet is low in fresh fruits and vegetables. The caloric content of potatoes is quite high: 100 g of tubers contain 70-83 kcal. Potatoes also contain biologically active substances - carotenoids, phenolic acids, flavonoids and glycoalkaloids (α-haconin and α-solanine), which in small quantities have anti-inflammatory, anti-allergenic and reducing cholesterol levels effects [5].
Growing potatoes does not require a special temperature regime, this crop is not picky about sunlight and humidity. The optimum growth temperature is 18-22°C, the minimum - 7°C, and the critical - 42°C. However, under conditions of excessive moisture, reduced soil looseness, and its permeability to moisture and air, the tubers are affected by phytopathogens and rot [6]. Reproduction of S. tuberosum is carried out both by seed and, most often, vegetatively. The advantage of the latter is that vegetative reproduction preserves the characteristics of the variety (productivity, taste, ripening period), but the disadvantage is that the seed material is subject to degradation due to its infection with phytopathogens [7]. For this reason, genetic improvement of potato varieties is currently a pressing objective.
However, there are currently numerous problems associated with growing potatoes, e.g. insufficient number of varieties adapted to growing conditions, low quality and efficiency of agricultural technologies, and inadequate conditions for storage, transportation and sale, all leading to spreading crop infections and pests [7]. Agrotechnological, chemical and biotechnological methods, and selection for disease resistance are widely used to control bacterial and fungal diseases of potatoes [8-10]. These methods have their advantages, as well as disadvantages: a) extra costs of chemicals and equipment for plant treatment, b) environmental pollution, c) prolonged (lasting 15-20 years) selection process, d) phytopathogens become resistant to chemicals and e) the contaminated potato plants lose resistance to the diseases [7, 11-14]. The limitations of chemical protection are that due to the uncontrolled use of pesticides and the rapid evolution of phytopathogens, chemicals quickly lose their activity [15-18].
Therefore, search for and development of alternative solutions to protect plants from pathogens, breeding new varieties of cultivated plants that would have steady resistance to a wide range of bacterial and fungal pathogens, elaboration of biotechnological and environmentally friendly methods for growing such plants all these tasks are urgent and important.
This review presents the characteristics of the main fungal and bacterial phytopathogens that affect potato plants; the analysis of the latest biotechnological approaches to the creation of potato plants resistant to a wide range of phytopathogens, in particular, the use of genetic engineering methods, including using TALEN and CRISPR-Cas9 genome editing technology, as well as the principles of marker-assisted selection.
2. THE MOST WIDELY SPREAD DISEASES OF POTATOES
During the growing season, potatoes are most often affected by late blight, alternariosis, rhizoctonia, bacteriosis and viral diseases. In 2014-2017, the most common potato diseases, for example in Ukraine, were late blight (infection of 6.1-91.3% of the potato growth area, the pathogen - Phytophthora infestans), alternariosis (infection of 16.1-91.5% of the potato growth area, the pathogen - Alternaria sp.), phomosis (1- 10% of areas, the pathogen - Phoma exigua), rhizoctonia (15-43.5%, the pathogen - Rhizoctonia solani), fusarium wilt (1-3%, the pathogens - Fusarium sp., Verticillium sp.), ring rot (23.7-94.8%, the pathogen - Clavibacter michiganensis subsp. sepedonicus), blackleg (2-88.3%, the pathogens - Pectobacterium sp., Dickeya sp) and viral diseases (0.7-37.2%) [19, 20].
2.1. Characteristics of Bacterial Pathogens in Potatoes
The most common and harmful species of phytopathogenic bacteria that infect potatoes are: Clavibacter michiganensis subsp. sepedonicus (the causative agent of potato ring rot) and Ralstonia solanacearum, which causes brown potato rot (Table 1). It should be noted that these bacterial phytopathogens are quarantined in Ukraine (On Amendments to the List of Regulated Pests, 2019).
| Phytopathogens | A Disease Caused by a Pathogen | Main Symptoms |
|---|---|---|
| Bacterial Phytopathogens | ||
|
Clavibacter michiganensis subsp. sepedonicus |
Ring rot of potatoes | The diagnostic sign is damage to the vascular system of stems and tubers. In the section, the affected areas are soft, have a lemon-yellow color and a homogeneous oily structure. The first signs of the disease appear at the end of flowering potatoes as a lesion of one or more shoots in the bush. The leaves of the affected plants are twisted and dried up |
| Ralstonia solanacearum | Brown rot of potato tubers | Necrotic brown spots are observed on the vascular bundles of the plant. Damaged leaves lose turgor and wither, and then dry up. When the stem of an infected plant is immersed in water, milky-white strands of bacteria are released from the stem fracture [25-29]. |
| Dickeya spp. | Black potato stalk | Yellowing of the lower leaves, blackening of the base of the stem and root system. |
| Fungal Phytopathogens | ||
| Phytophthora infestans | Late blight | Disease symptoms appear on plants in 2-5 days after contamination: brown moist necrotic spots are formed on the leaves, on which a white plaque appears in the wet period [34, 35]. |
| Fusaruim sp. | Dry rot of potatoes | Grayish-brown wrinkled spots appear on the surface of potato tubers. Convex pink or grayish-white pads are formed on the surface of such spots. |
C. michiganensis is a biotrophic pathogen that can infect a wide range of hosts. Currently, nine subspecies of C. michiganensis are known to infect cereals, legumes and nightshades. The causative agent of ring rot in potatoes is a subspecies of C. michiganensis subsp. sepedonicus [21, 22]. It is an immobile non-spore-bearing gram-positive rod-shaped bacterium with a cell size of 0.5-1.0 - 0.4-0.6 μm [22, 23]. The natural reservoirs of C. michiganensis are weeds such as Solanum nigrum, S. douglasii, S. trifolium, Datura stramonium, Chenopodium album and Amaranthus retroflexus. Sources of infection - infected seeds and plant debris. The pathogen affects the vascular system and spreads rapidly to all tissues and organs of the plant [22-24].
Infection of potatoes with C. michiganensis usually occurs at high temperatures (25-27°C) and humidity (80-85%). The accumulation of the pathogen in the soil is facilitated by monocultures, and the source of damage is undigested plant remains. The difficulty of controlling this phytopathogen is due to the long latent period of disease development and a wide range of hosts belonging to the Solanaceae family, which complicates crop rotation [22].
An extremely dangerous and harmful bacterial phytopathogen that infects plants of the nightshade family is R. solanacearum, which causes the brown rot of potato tubers. Although no outbreaks of R. solanacearum have been detected in Ukraine over the past 15 years, the active import of food and seeds from European countries and the favorable climatic conditions in Ukraine pose a risk of the introduction of this pathogen [25].
R. solanacearum is a biotrophic plant pathogen, gram-negative bacterium with rod-shaped cells 0.5-1.5 μm in size, and does not form spores [25]. This bacterium is classified as quarantine phytopathological microorganisms in Ukraine. The source of infection is usually plant remains, infection occurs through damaged roots. Natural reservoirs of infection are weeds Amaranthus spinosus, Chenopodium album, Euphorbia hirta, Malva sp., Solanum nigrum, S. dulcamara, Vicia sp., etc [25-29]. Depending on the hosts, there are five races R. solanacearum, of which infect mainly plants belonging to the Solanaceae family, as well as banana trees, ginger and mulberry. As a result of phylogenetic analysis, 4 phylotypes and 6 biovars of R. solanacearum were described: Asian phylotype I (which includes biovars 3, 4, 5), American phylotype II (biovars 1, 2, 2T and races 2, 3), African phylotype III, biovars 1 and 2T), and Indonesian phylotype IV (biotype 1, 2, 2T) [27].
Outbreaks of plant diseases caused by R. solanacearum race 3 have been reported in a number of European countries (Belgium, Germany, Britain, the Netherlands, France, Italy, Spain, Greece) [26-29].
The success of the control of diseases caused by R. solanacearum depends on the timely and accurate diagnosis of diseases. Serial dilution methods, enzyme-linked immunosorbent assay and PCR diagnostics are used for diagnosis. The difficulty of controlling this pathogen is due to its ability to survive adverse conditions for several months in deep soil layers (up to 30 cm), water bodies, plant remains, weed vascular systems, long latency and infection of a wide range of hosts. The success of controlling the diseases caused by R. solanacearum depends on their timely and accurate diagnostics. To identify the pathogen, serial dilution methods, enzyme-linked immunosorbent assay and PCR diagnostics have been used. The difficulty of controlling this pathogen is due to its ability to survive prolonged, up to several months, adverse conditions in deep, down to 30 cm, soil, water bodies, plant remains, weed vascular systems, long latency periods and infecting of a wide range of hosts [28, 29].
No less dangerous is one of the causative agents of blackleg potatoes - bacteria of the genus Dickeya. Strains of Dickeya spp. cause significant economic losses in potato potatoes that are grown in Western Europe. The most harmful are D. dadanthii and D. zeae, which affect potato plants in hot climates, and D. dianticola, which is adapted to temperate climates [30].
Diagnosis and control of bacterial diseases in potatoes are also complicated by the fact that bacterial pathogens are usually mutually tolerant - that is, strains of different species of bacterial pathogens (X. campestris, P. syringae, C. michiganensis, R. solanacearum) do not compete with each other for food and do not affect each other's growth. Thus, infected potato plants can be simultaneously colonized by several pathogens [28, 29, 31].
2.2. Characteristics of Fungal Pathogens of Potatoes
Among the fungal pathogens of plants, special attention is drawn to Phytophthora infestans (Mont.) Be Bary - an extremely harmful pathogen that has caused large-scale damage to potato crops since the mid-19th century, against which there is still no effective means of control [32, 33].
P. infestans is a heterothallic hemibiotrophic oomycete that damages all terrestrial and underground plant organs [34, 35]. There are several alternative sources of infection and natural reservoirs of infection:
- Oospores, which are formed as a result of sexual intercourse on infected plant remains. At the stage of oospores, P. infestans can remain viable in the soil for several months and even years;
- Mycelium in infected potato tubers. On infected shoots, which germinate from infected tubers, sporulation begins quickly.
- alternative carriers, usually members of the family Solanaceae - Solanum melongena, S. incanum, S. indicum, Datura stramonium, D. metel, Hypomea sp., Lycium hamilifolium, Nicotiana glauca, Petunia sp., Physalis angulata,etc [32]. Infection occurs via the following mechanism. When the sporangium falls on the leaf, motile flagellar zoospores get released. The latter settle on the leaf surface, incise and germinate in the germ tube, which forms appressoria and penetrates the leaf tissues through the stomata or through the cells' epidermis. At an optimum temperature of 13°C infections occur within 3-4 hours. Hyphae form haustoria inside the leaf, in the intercellular space, due to which the biotrophic type of nutrition occurs. The first necrotic spots appear within 2-5 days at a favourable temperature of 22-24°C. After a successful infection takes place, a large number of lemon-shaped sporangia (24,000/cm2) appear on the surface of the infected tissues. Spore-laying occurs at temperatures from +3°C to +26°C and with 90% humidity. Sporangia are rapidly carried by wind and water and infect new plants [32, 34, 35].
The presence of a latent biotrophic phase of late blight complicates the diagnosis and control of the disease. When the first necrotic spots appear, plant treatment with fungicides will have no effect on the development of the disease.
Compared to other stramenopiles, P. infestans has a large genome of 240 million bp. with a lot of repeating sequences. It is the genetic lability of late blight, that allows it to effectively overcome the protective strategies of plants. It happens due to such features of the pathogen as 1) the presence of large numbers of non-coding RNA, 2) expression of pseudogenes, 3) the presence of large untranslated regions of the genome (UTRs) 4) due to these regions, expression of genes takes place via the mechanism of alternative splicing 5) the presence of a large number of single nucleotide polymorphisms (SNP) [36].
Other dangerous phytopathogens that infect Solanum tuberosum are members of the family Fusaruim sp. (Table 1 and Fig. 1). Dry fusarium rot of potato tubers is a disease of storage. The causative agents of dry fusarium rot of potatoes are most often F. sambucinum, F. solani, F. oxysporum. Dry potato rot is spread everywhere where this crop is grown, and the harmfulness of this disease is no less than late blight. Seed material affected by fusarium rot and planted in the soil causes rarefaction of plantings and shortage of yields. Affected tubers delay the growth and development of plants during the vegetation season and premature wilting. Loss of tubers from dry fusarium-caused rot during storage is 7-23%, and in the case of high humidity - up to 60% [32, 37].
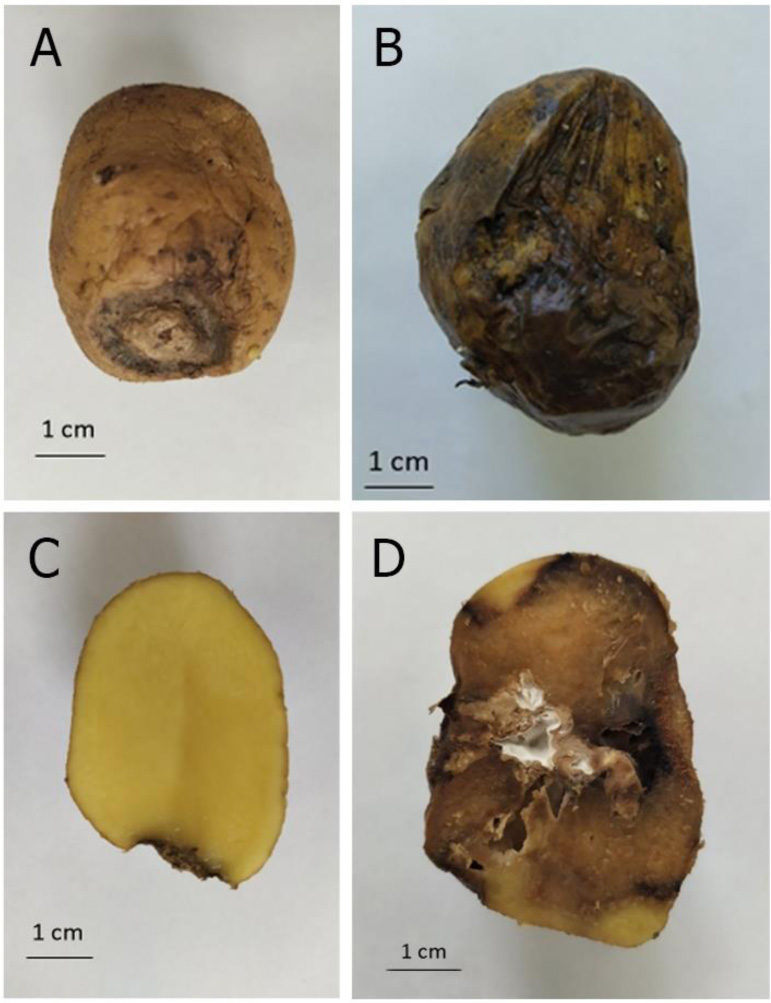
Dry potato rot not only reduces the yield but also contaminates the tubers with mycotoxins that have cyto-, geno-, neuro- and hepatotoxic effects on animals and humans [38]. Among the toxins produced by F. sambucinum, the main ones are thiotecenes (15-monoacetoxyspirpenol and 4,15-diacetoxyspirpenol), sambutoxin and eniatin [39].
Fusarium species are soil pathogens that infect the vascular system of host plants. The life cycle of Fusarium species consists of several distinguished stages, such as dormancy, invasion and the final, saprotrophic stage, during which the sporulation of the pathogen is formed. Thus, Fusarium species are saprotrophic or necrotrophic pathogens. During the dormant stage, all life forms of the fungus (mycelium, chlamydospores, micro- and macroconidia) are inhibited by mycostasis. During the invasion stage, the pathogen penetrates into the roots, colonizes the endoderm and root cortex, moves to the xylem of the stem and leaves, and develops the disease symptoms (wilting, growth retardation) and early death of the plant. During the saprotrophic stage, sporulation is formed. At this stage, the fungus tolerates adverse conditions. Germination of spores is stimulated by root exudates of the host plant or contact with fresh non-colonized plant remains. After successful colonization of the host vascular system, microconidia are formed inside the xylem on the mycelium. Then, they are transferred by the vascular system, germinate and form new hyphae which result in the amplification of the symptoms. In the final stage of the disease, the death of the host plant takes place [39].
The life cycle of Fusarium spp. consists of sexual and asexual stages. As with most fungi, Fusarium species are haploid for most of their life cycle. Haploid mycelium is formed during both stages. During the asexual stage, as a result of apomixis, mycelial structures form 3 types of mitotic spores: microconidia in conidiophores, macroconidia in sporodochia and chlamydospores on hyphae and macroconidia. In the life cycle of Fusarium spp. sexual and asexual stages are present. As with most fungi, Fusarium species are haploid for most of their life cycle. Haploid mycelium is formed during both stages. During the asexual stage, as a result of apomixis, mycelial structures form 3 types of mitotic spores: microconidia in conidiophores, macroconidia in sporodochia and chlamydospores on hyphae and macroconidia [40].
Sexual reproduction begins with the formation of hyphae with dinuclear cells (the dinuclear phase is typical of Ascomycota). F. sambucinum is a heterothallic bisexual species, so the formation of ascospores requires contact of the mycelium of two different sexes. The dinuclear cells of F. sambucinum form initial cells, from which fruit bodies are further formed - cup-shaped perithecia filled with asci. The latter are represented by tubular sacs filled with ascospores formed as a result of meiosis. Ascospores are released from the asci through the mouth of the peritoneum. During the sexual stage, F. sambucinum tolerates adverse conditions during the winter period [40-42].
F. sambucinum forms spindle-shaped elliptically curved macroconidia in aerial mycelium, pyonotes and occasionally in sporodochia. Conidia have 5, rarely 3 membranes. Spores are of pink-orange color. Aerial mycelium can be white, pale red or pinkish, very fluffy or dense. Stroma can be yellow or bright red.
In most cases in Ukraine, tuber fusarium causes a species of F. sambucinum, which was found in almost 50% of the analyzed samples. The most effective method of controlling dry Fusarium rot of potato tubers is to create resistant varieties, but currently, there are no varieties of potatoes with high resistance to this disease, which may be due to the mixed species composition of Fusarium spp., which colonize individual plants [21]. Thus, according to another study [20], the resistance of 173 potato varieties to dry Fusarium rot was analyzed, and among the analyzed varieties, no single one with at least a relatively high resistance to this disease was found.
3. STRATEGIES FOR THE DEVELOPMENT OF POTATO RESISTANCE TO PHYTOPATHOGENS
Promising methods of plant disease control are biological methods, in particular: 1) contamination with avirulent strains of phytopathogens, 2) treatment of crops with drugs based on antagonistic microorganisms that activate the immune response of plants by ISR (induced systemic resistance), such as Pseudomonas sp. Streptomyces sp., Bacillus sp. The advantages of biological control methods are longevity and the ability to self-sustain, self-repair and spread. The disadvantage of such methods is the unpredictable impact on the chemical and microbiological composition of soils [10, 28, 43, 44].
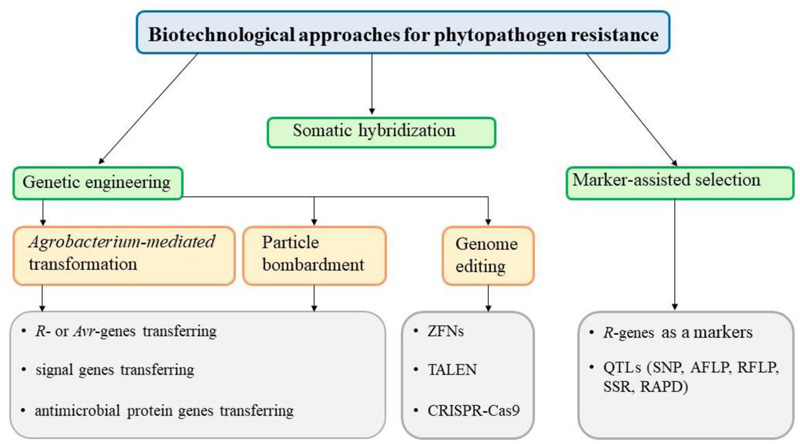
An alternative method of plant protection is the use of biotechnological methods (Fig. 2), including somatic hybridization, marker-assisted breeding, genetic engineering, and genome editing technologies. Methods of genetic engineering enable the transfer of genes of resistance to phytopathogens into the genomes of valuable plant varieties.
According to the mechanisms of resistance to phytopathogens, three groups of target genes can be distinguished: 1) genes of resistance and avirulence (R- and Avr-genes, respectively), 2) genes of resistance to bacterial phytotoxins, 3) antimicrobial peptides of plant, animal or bacterial origin that reveal bactericidal activity [10, 45-54].
3.1. Use of Resistance and Avirulence Genes to Generate Potatoes Resistant to Phytopathogens by Genetic Engineering
One of the ways to increase the resistance of potato plants to phytopathogens is to introduce resistance genes (R-genes) isolated from wild related species by methods of genetic transformation. The degree of resistance formation depends on the choice of R-genes and the diversity of Avr pathogen genes. In particular, the avirulence gene Avr-vnt1 P. infestans and the R-gene Rpi-vnt1.1 of potatoes are genetic components of the “gene-gene” interaction, which ensures the resistance of host plants to this pathogen. It is shown that the transfer of Rpi-vnt1.1 gene to Desiree potato plants from Solanum venturi by Agrobacterium-transformation contributed to the increase of resistance of these plants to P. infestans, which was followed by low expression of the corresponding avirulence gene Avr-vnt1 of two isolates of EC-1 P. infestans [55, 56]. In the study [14], the Rpi-mcq1 gene from S. mochiquence was isolated and transferred to the Desiree potato cultivar. The obtained lines had been field studied for 3 years (2010-2012), which showed that transgenic potato lines with the gene Rpi-vnt1.1 were resistant to P. infestans.
To date, quite a wide range of R-genes of wild species related to S. tuberosum, which are able to induce resistance to P. infestans, have been cloned and transferred to potato cultivars. Successful transfer of both single and several genes simultaneously has been performed. Examples of such genes are R1, R2 and R3a, R3b isolated from S. demissum; Rpi-blb1 (RB), Rpi-blb2, Rpi-blb3 with S. bulbocastanum; Rpi-vnt1.1 and Rpi-vnt1.2 from S.venturii; Rpi-mcq 1 of S. mochiquense [11, 57], etc. Fortuna potato plants with two R-genes (Rpi-blb1 and Rpi-blb2) isolated from the systematically related species S. bulbocastanum were obtained by the European BASF company. The resulting plants showed resistance to late blight, but this variety has not been presented on the market [11].
In addition, to ensure resistance to late blight, it was proposed to create potato varieties polymorphic to R-genes [13]. The potato varieties polymorphic to R-genes can be created by a simpler and more efficient method of cysgenesis. It allows the transfer of genes of useful traits from the genomes of wild Solanum species to the genomes of industrially valuable potato varieties by genetic transformation [13]. Attempts to obtain phytophthora-resistant potato plants simultaneously with three Rpi genes, Rpi-blb1and Rpi-blb2 isolated from S. bulbocastanum, and Rpi-vnt1.1 isolated from S. venturii, by cysgenesis were quite successful [58].
However, resistance to late blight induced by overexpression of R-genes has a number of disadvantages. In particular, such resistance is not stable, it can be lost due to the variability of the virulence of the pathogen, in particular P. infestans. Therefore, the idea to use the products of avirulence genes as effectors that can cause indirect hypersensitivity and disease resistance in plants was suggested. In particular, it has been shown that transient expression of the avirulent genes P. infestans PiAvr1, PiAvr2, PiAvr3aKI, PiAvr4 and PiAvr8 in potato plants can cause a hypersensitive response to P. infestans infection, expression of the corresponding R genes and, consequently, resistance to this pathogen [59].
A very promising way to obtain plants resistant to pathogens is to induce their own immune response, for example, by pathogen-targeted immunity. It was found that potato plants have a number of receptor proteins for pathogen recognition. Thus, receptor-like ELR proteins have been identified which, via binding to BAK1 / SERK3 co-receptors, recognize elisitin pathogens, in particular Phytophthora species. They induce a plant immune response to infection with pathogens of this genus [60]. It has been shown that the isolation of genes encoding the synthesis of ELR proteins in wild related species, and their transfer to the genome of potato cultivated plants, gives such plants resistance to late blight. Thus, the gene of receptor-like protein ELR isolated from the wild species Solanum microdontum and transferred to potato cultivars provided the formation of their resistance to P. infestans [61].
Another strategy for creating plants resistant to a wide range of pathogens is the overexpression of regulatory genes involved in signaling cascades of stress responses, which can provide stable resistance, including to pathogens. G-proteins play a key role in the development of plant disease resistance. Thus, stable overexpression of the AtRop1 gene (DN-AtRop1), which encodes one of the G-proteins, contributed to the increased resistance of potato plants to P. infestans [62]. In addition, it has been shown that genes encoding signaling proteins isolated from some plant species can activate the immune response in other species. Thus, the transfer of the lectin receptor kinase gene LecRK-I.9 Arabidopsis thaliana to S. tuberosum plants increased the resistance of these plants to P. infestans [63].
In addition, potato plants resistant to P. infestans were obtained using host induced gene silencing (HIGS). Host-induced gene silencing (HIGS) is used to determine the function of pathogen genes in plants. In HIGS, double-stranded RNA (dsRNA) homologous to pathogen genes can be transferred to plant cells, causing pathogen gene silencing [64]. The main goal of this strategy is to achieve longer stability than when using R-genes [11].
An important source of genes for resistance to phytopathogens is wild species of nightshade. More than 40 resistance genes have been identified in wild species of Solanaceae, about 20 of which have been transferred to cultivars. Among wild species, most resistance genes have been identified in S. chilense, S. peruvianum, S. habrahaides and S. pimpinellifolium [65]. Thus, in tomato plants, the Ve locus was identified, which contains 2 inverted Ve1 and Ve2 genes that provide resistance to Verticillium species. These genes encode glycoproteins located on the cell surface and contain signal sequences of receptor-mediated endocytosis. The tomato receptor protein gene Ve1, which is able to recognize Ave1 protein V. dahliae when expressed in potato plants, provided resistance to V. albo-atrum [66].
3.2. Use of Antimicrobial Peptide Genes to Create Plants Resistant to Pathogens
Another area of the creation of plants resistant to phytopathogens by methods of genetic engineering is the introduction into the plant genome of genes of antimicrobial peptides of plant, animal, or bacterial origin, which have bactericidal activity. Such peptides may be PR (pathogenesis-related) plant proteins [67, 68]; α-chain cecropines of Lepidoptera, magains of tropical amphibians, β-folded defensins or protegrin, linear melittin from bee venom [69, 70]; genes of animal peptides - magainins, cercopins, defensins, chitinases, lactoferrin [71], defensin and bombinin from leukocytes and epithelial cells of rabbits, and lactoferrin from cow's milk [67]; bacterial peptides (gramicidin from a soil strain of bacteria of the genus Bacillus sp. (Hotchkiss and Dubos in 1940).
3.2.1. Antimicrobial Peptides of Plant Origin
PR proteins are small peptides with a molecular weight of 5-75 kDa, which were assigned to 17 families according to their activity. Examples of some PR proteins are chitinase, glucanase, thaumatin-like proteins (TLPs), proteinase inhibitors, peroxidases, ribonuclease-like proteins (RLPs), defensins, thionines, lipid transporter proteins (LTPs), oxalate oxidase (OXOs), etc [8, 9, 68, 72]. Genes of plant hydrolytic enzymes β-glucanases and chitinases belonging to the PR-2 and PR-3 families, respectively, are widely used to increase resistance to phytopathogens. These enzymes destroy the cell wall, which prevents the growth of hyphae and their penetration into plant tissues [60, 71, 73].
Another antimicrobial peptide of plant origin is the cysteine-rich protein Snakin-1 (SN1), which has antibacterial activity against a wide range of pathogens. This protein was isolated from potato tubers, variants of the gene encoding this protein were cloned, and the ability to induce plant resistance to phytopathogens was shown. Thus, potato plants (S. tuberosum subsp. tuberosum cv. Kennebec), which were transformed with a vector with the SH-1 protein gene, showed resistance to phytopathogens such as Rhizoctonia solani and Erwinia carotovora [74].
3.2.2. Antimicrobial Peptides of animal Origin
Since plant pathogens are able to overcome the mechanism of “gene-against-gene” resistance, an effective strategy to increase the resistance of cultivated plants to phytopathogens is the transformation of plants with genes encoding non-plant AMP that have antimicrobial activity against many pathogens [69, 70, 75, 76]. Examples of such proteins are α-chain cecropines of Lepidoptera, magainins of tropical amphibians, β-folded defensins or protegrin, linear melittin from bee venom, and the like. However, some AMPs may be toxic to potential consumers, such as cercopins having hemolytic activity [69, 70]. Lactoferrin is a potentially safe antimicrobial protein for human health. Due to its numerous beneficial properties, lactoferrin is a promising gene of interest for expression in the genomes of cultivated plants [75, 77]. The use of the lactoferrin gene to increase the resistance of transgenic plants to disease may increase the effectiveness of the natural defense mechanisms of the plants by binding iron and by directly destroying pathogen cells. The antibacterial activity of lactoferrin and oligopeptides derived from lactoferrin has been confirmed against a large number of bacterial pathogens in humans, animals and plants [51-53, 78].
As reported [79], the transfer of the human lactoferrin gene into the potato plant genome makes it possible to obtain lines resistant to phytopathogenic bacteria R. solanacerum, C. michiganensis subsp. seredonicus, as well as the fungal pathogen P. infestans. In addition, several studies have reported the effectiveness of external treatment of plants with lactoferrin to protect against disease, and this approach was more effective than the use of contact fungicides [80-83]. (Table 2).
It should be noted that in addition to genetic engineering, another method of transferring traits of resistance to various pathogens from one plant species to another, and to potatoes in particular, is the method of somatic hybridization (protoplast fusion method). Given the considerable evolutionary diversity of wild relatives of the potato and other related Solanum species [84-86], which are the source of important genes, they are of great interest for potato quality improvement using various biotechnological methods, including cell fusion methods.
A comprehensive review on the use of somatic hybridization to enhance the potato gene pool by introducing genes from wild species has recently been published, clearly describing advances over the past 40 years in transferring pathogen resistance traits (including bacterial and fungal disease resistance traits) from various Solanum species into potatoes [87].
3.3. Marker-assisted Selection to Improve Potato Traits
One of the most significant achievements of modern molecular genetics and biotechnology is the decoding of the genomes of various organisms, including potatoes, and the creation of genetic maps. By identifying qualitative traits that contain genes responsible for morphological traits, plant development and tuber quality, as well as potato resistance genes, it is possible to perform marker-associated selection that is much less time-consuming and more accurate than classical selection. However, even having this approach, the selection is a complex process, as most industrially valuable traits are polygenic and are significantly influenced by environmental factors; also, the selection is complicated by the fact that the species S. tuberosum is tetraploid (2n = 4x = 48), so it is difficult to achieve homozygosity for a certain trait [12].
Selection for resistance to phytopathogens is delivered using marker-associated approaches, cultivation in the laboratory, field and greenhouse conditions [88]. However, although modern advances in molecular biology have greatly expanded the possibilities and facilitated the selection process, the latter on some traits still remains a time-consuming process [88].
| Genes | Sources | Method of Transformation | Resistence to Pathogens | Potato Cultivar | References | |
|---|---|---|---|---|---|---|
| Transfer of Resistance Genes as Target Genes | ||||||
| Rpi-vnt1.1. | S. venturi | Agrobacterium-mediated | Phytophtora infestans | Desiree | [55, 56] | |
| Rpi-mcq1 | S. mochiquence | Agrobacterium-mediated | P. infestans | Desiree | [14] | |
| R1, R2, R3a, R3b | S. demissum | Agrobacterium-mediated | P. infestans | Desiree | [11, 57] | |
| Rpi-blb1 (RB), Rpi-blb2, Rpi-blb3 | S. bulbocastanum | Agrobacterium-mediated | P. infestans | Desiree, Fortuna | [11, 57] | |
| Rpi-vnt1.1, Rpi-vnt1.2 | S.venturii | Agrobacterium-mediated | P. infestans | Desiree | [11, 57] | |
| Transfer of Avirulence Genes as Target Genes | ||||||
| PiAvr1, PiAvr2, PiAvr3aKI, PiAvr4, PiAvr8 | P. infestans | transient expression, Agrobacterium-mediated | P. infestans | Qingshu9; Longshu7 | [59] | |
| Gene transfer of receptor-like proteins recognizing pathogens and signaling proteins | ||||||
| Gene of receptor-like protein ELR | S. microdontum | Agrobacterium-mediated | P. infestans | Desiree | [61] | |
| AtRop1 | A. thaliana | Agrobacterium-mediated | P. infestans | Shepody | [62] | |
| LecRK-I.9 | A. thaliana | Agrobacterium-mediated | P. infestans | Desiree | [63] | |
| Ve1, Ve2 | L. esculentum | Agrobacterium-mediated | Verticillium albo-atrum | Desiree | [66] | |
| Gene Transfer of Antimicrobial Peptides | ||||||
| Snakin-1 (SN1) | S. tuberosum | Agrobacterium-mediated | Rhizoctonia solani та Erwinia carotovora | Kennebec | [74] | |
| hLf (human lactoferrin) | Human | Agrobacterium-mediated | Ralstonia solanacerum, Clavibacter michiganensis subsp. seredonicus, P. infestans | Vernisage, Levada, Svitanok Kyivskyi, Zarevo | [79] | |
| Application of genome editing technology | ||||||
| StDND1 та StCHL1 (S-genes) | S. tuberosum | Agrobacterium-mediated, CRISPR/Cas9 | P. infestans | Désirée and King Edward | [83] | |
The difficulty in the selection of valuable potato varieties for resistance to various phytopathogens is rooted in the polygenic nature of resistance and the coupling of resistance genes with undesirable traits [13, 29, 31, 89]. Since crossbreeding with wild species of the genus Solanum may transfer, additionally to target traits, clutched non-target traits, and a reverse crossbreeding with the parent variety is required to remove the pool of linked genes, the selection process can last for decades. However, no genes for resistance to certain phytopathogens, such as C. michiganensis and R. solanacearum, have been identified among wild species of the Solanaceae family. However, some wild species of Solanaceae, which are able to provide tolerance to this bacterium, are widely used in breeding [89].
The search for molecular markers that could be used to select plants for resistance to pathogens remains quite topical. In the period from 2006 to 2015, the Wageningen University and Research Center in the Netherlands developed an investigation aimed at finding genes or clusters of genes that could be used for marker-assisted breeding for resistance of potato plants to late blight [61]. As a result of this work, a number of potato varieties resistant to late blight have been created. To achieve that, 13 R-genes isolated from wild-related species were used as markers, but to enter the market, a number of additional studies and backcrosses are still required [57].
Using marker-assisted selection and various marker systens such as AFLP, RFLP, SSR, RAPD, or genes for qualitative traits (QTLs) is considered promising for the development of pathogen-resistant plants, including potatoes [90].
3.4. Genome Editing Methods as a Promising Approach to Increasing Resistance to Phytopathogens
The latest trends in plant biotechnology are based on avoiding introducing alien DNA into their genome but are rather aiming at applying the genome editing technology, including the method of “zinc fingers” (zinc finger nucleases, ZFN), at using transcription activator-like effector nucleases (TALENs), the clustered regulatory interspaced palindromic repeats or CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) [91, 92]. Recently, the greatest preference in biotechnology is given to the use of CRISPR technology - a natural system of immune response of bacteria to the invasion of bacteriophages and foreign genetic fragments [93]. The CRISPR-Cas method is based on the use of short single-guide RNA (sgRNA), which is complementary to the target region of the genome that needs editing, and Cas9 nuclease [92]. Such a system can quite accurately induce mutations in the target region due to sgRNA and the ability of nuclease (Cas9 or other) to perform a double-strand break in this region, which can then be regenerated by inhomologous connection of ends or homologous recombination [92].
An attempt to induce targeted mutagenesis in potato plants using CRISPR-Cas9 and TALEN technology proved to be quite effective, which led to the knocking out of target genes [94-96]. Attempts to obtain potato plants resistant to low temperatures and herbicides, plants with altered starch quality, and reduction of enzymatic darkening of potatoes have been rather successful [97, 98].
Obtaining disease-resistant potato plants is also possible with the use of genome editing technologies [83, 99]. In particular, potato plants with increased resistance to late blight were obtained using CRISPR / Cas9 technology by mutagenesis (silencing) the sensitivity genes (S-genes) StDND1 and StCHL1 [83]. Although genome editing technologies such as CRISPR / Cas9 or TALEN are increasingly used to improve the properties of crops [100], in particular, potatoes [101, 102], the application of genetic engineering approaches to improve the properties of valuable potato varieties is still relevant because it allows the direct transfer of genes of important traits to the genomes of valuable varieties. It is also important to note that the risks associated with using genetically modified potatoes are low: the rate of transgene release into the environment by pollen or seed transmission is relatively low at 6 out of 27 [103], resulting from low sexual compatibility with wild species such as Solanum nigrum. or S. dulcamara [11, 12].
CONCLUSION
Given the significance of potatoes as an agricultural crop, it is important to create S. tuberosum lines resistant to dangerous phytopathogens of bacterial nature, such as R. solanacearum (which causes brown rot of tubers) and C. michiganensis subsp. sepedonicus (the causative agent of ring rot of potato tubers), as well as to such aggressive phytopathogenic fungi as P. infestans (causative agent of late blight) and F. sambucinum (causative agent of dry rot of potato tubers). One of the promising approaches to increasing the resistance of cultivated plants to these phytopathogens is the use of genetic engineering methods, marker-assisted selection and genome editing methods.
AUTHOR'S CONTRIBUTIONS
Olena Kvasko: Data analysis, writing manuscript; Yuliia Kolomiiets: Data analysis, critical remarks; Anastasiia Buziashvili: Data analysis, writing, figure drawing; Alla Yemets: Conceptualization, data analysis, review, critical remarks.
LIST OF ABBREVIATIONS
| UTRs | = Untranslated Regions Of The Genome |
| SNP | = Single Nucleotide Polymorphisms |
| dsRNA | = Double-Stranded RNA |
| HIGS | = Host Induced Gene Silencing |
| SN1 | = Snakin-1 |
| QTLs | = Qualitative Traits |
| TALENs | = Transcription Activator-Like Effector Nucleases |
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was supported partially by a grant from the Ministry of Education and Science of Ukraine (“Induced resistance and control of phytopathogenic bacteria in novel biotechnologies for growing vegetable crops using growth stimulants with elicitor activity” (0120U102106, 2020-2023).
CONFLICT OF INTEREST
Dr. Alla Yemetsis the Editorial Advisory Board of the journal The Open Agriculture Journal.
ACKNOWLEDGEMENTS
The authors are grateful to Dr. V. Kyrylenko (Institute of Food Biotechnology and Genomics, National Academy of Sciences of Ukraine, Kyiv) for the final improvement of the English version of the manuscript.


