All published articles of this journal are available on ScienceDirect.
Bacteriological Quality and Antibiotics' Susceptibility Profile of Small-medium Scale Commercial Fish farms in Nigeria
Abstract
Background:
Fish currently provide 6.7% of all proteins consumed by humans globally; nevertheless, the aquaculture system has been linked to fish, environmental contamination and disease outbreak. The aim of this study was to determine the bacteriological quality and the antibiotic resistance profile of bacteria from water samples of pond stocked with Tilapia and Catfish in Ile-Ife, Osun State, Nigeria.
Objective:
To isolate, identify and characterise heterotrophic bacteria and test for the antibiogram of detected Coliforms.
Methods:
Water samples were collected from ponds stocked with Tilapia and Catfish, and tested for total heterotrophic and coliform bacteria as well as the antibiogram. The susceptibility of the isolates was tested using the Kirby-Bauer disc diffusion method on Mueller Hinton agar.
Results:
A total of 40 isolates were recovered from the water samples, of which 5 species were Gram positive bacteria representing two genera, and 35 species were Gram negative bacteria representing four genera. The temperature for all ponds ranged from 25°C to 28°C. The mean bacterial count varied from 1.9×104 to 5.4×104 CFU/ml per fish pond. All isolates were 100% resistant to ceftazidime, cefuroxime and augmentin. More resistance to cefixime (80%) and gentamicin (73.3%) and nitrofurantoin (66.7%) was also recorded. However, only 16.6% and 8.3% of the isolates were resistant to ciprofloxacin and ofloxacin, respectively. The multiple antimicrobial resistance index (MARI) ranged from 0.5 to 0.9. The water physicochemical parameters (temperature and pH) and the type of bacteria detected in all pond types did not differ significantly.
Conclusion:
Fish pond is a reservoir of multi-drug resistant bacteria that could serve as environmental source of drug resistance gene transfer. This calls for effective monitoring and assessment as well as management devices for the protection of community and environmental health.
1. INTRODUCTION
Aquaculture fish production in Nigeria has grown from 0.1% to 0.4% from 1995 to 2016 [1]. The FAO estimates an increase in fish production in Nigeria by 2030 to about 18.2%, while export will increase by about 6.6% [1]. These figures show the importance of fish to nutrition and food security. However, recent reports on fish meat safety have heightened consumers' fear regarding fish meat and fish products. For instance, cases of fish-borne disease caused by pathogenic microorganisms like Salmonella, Listeria monocytogenes and Vibros spp. have been reported in several parts of the world [2, 3].
Often the primary driver of such food intoxication is the farming environment and feed given to the fish. Since fish lives in water, the quality of water directly impacts fish productivity, fish products, human and environmental health. Water quality is one of the most overlooked aspects of pond management until it adversely affects the quality of fish production. The factors which influence the use of water for fish culture include dissolved oxygen, pH, hardness, turbidity, alkalinity, ammonia and temperature. The level of pollution of a given water body is indicated by other parameters such as biological oxygen demand and chemical oxygen demand [4, 5].
Fishes are reared in different water culture media or confinement such as concrete, earthen or plastics ponds. Concrete and earthen ponds have been the widely used culture system for fish. Earthen pond system of fish cultivation has been the most established method of fish culture in Nigeria. Fishes reared in these environments are contaminated by both pathogenic and opportunistic microorganisms. The contamination of these culture systems has been attributed to poor water quality, high stocking densities and the use of animal manure and contaminated feed [6, 5]
Due to the high cost of feeding, farmers use animal manure to supplement feeding. The use of organic manure also leads to the release of high concentration of opportunistic and pathogenic microorganisms into the ponds, which pose a threat not only to fish health but also to the environment [7]. Also, these microorganisms in fish and fish ponds portend grave consequences for public health [2, 3]. Some of these microorganisms possess resistant determinant, which enhances their potential for infecting consumers. Such resistant pathogens are capable of undermining effective health outcomes and prolonging hospitalisation of patients. Hence, it is essential to document the microflora and antimicrobial resistance associated with the fish environment since the microbial flora of a cultivated fish is an expression of its aqueous environment.
2. MATERIALS AND METHODS
2.1. Study Area
The study area is Fajuyin and Oke Opa within Ile-Ife, Osun State in Nigeria, which lies between a latitude of 70˚N 50’N and longitude of 4˚ 69’E (Fig. 1). The climate is tropical, and mix farming system is conventional. Farmers combine fish production, livestock production, and plantain and banana plantation. Hence, having a combination of concrete, plastic and earthen fish ponds stocked with Tilapia and Catfish is more profitable. All ponds surveyed in the study were privately owned. The Earthen ponds and plastic ponds were covered with a net to prevent birds and predatory animals from accessing the pond, while the concrete ponds were in a fenced area and then roofed.
2.2. Sample Collection, Temperature and pH Measurement
Water samples were aseptically collected from the ponds (from April to July, 2018) using sterile screw-capped kegs. Composites samples were obtained by collecting at different sampling points and depths of 30cm below the water surface from two separate concrete ponds, earthen ponds, and plastic ponds, respectively (Table 1), which makes a total of 6 samples. The water samples were transported in a box containing ice packs to the laboratory for microbiological and biophysical analysis. The temperature of water samples was measured at the sites of sampling with a standard laboratory mercury thermometer (Assutech, South Africa). The pH of water samples was determined using pH meter (designer water, South Africa) after its calibration.
Table 1.
| Sampling Stations | Observed Features |
|---|---|
| C1(Concrete pond 1) | The pond is at Fajuyi area in Ile-Ife. The pond is in a closed, fenced and roofed area; also refuse is being dumped behind the fence and waste water from bathroom flows close to the concrete pond. |
| C2(Concrete pond 2) | The pond is at Fajuyi area in Ile-Ife. The pond is in a closed, fenced and roofed area; also refuse is being dumped behind the fence and waste water from bathroom flows close to the concrete pond. |
| E1(Earthen pond 1) | The pond is at Fajuyi area in Ile-Ife. The station is close to a primary school and a farm land, surrounded with plantain and banana plantation with other trees; and the pond was covered with a net to prevent bigger animals from picking up fish. |
| E2(Earthen pond 2) | The pond is at the Fajuyi area in Ile-Ife. The station is in a compound, with a well close to it, surrounded with plantain and banana plantation with other trees, and the pond was covered with a net to prevent bigger animals from picking up fish. |
| T1(Tank pond 1) | The pond is at Oke Opa area in Ile-Ife. The pond is in a compound where snails and poultry are being reared, having a cashew tree in the compound. The pond is stocked with African cat fish covered with net. It also has a running tap in the compound which is used for fish farming. |
| T2(Tank pond 2) | The pond is at Oke Opa area in Ile-Ife. The pond is in a compound where snails and poultry are being reared, and a cashew tree is also present. The pond is stocked with African cat fish covered with a net. It also has a running tap in the compound which is used for the fish farming. |
2.3. Isolation of Total Heterotrophic Bacteria
Sterile test tubes containing 9ml sterilised water were used for each sample serial dilution. Up to 1ml of each homogenised sample was seeded into the first test-tube using a sterile syringe, and swirled gently to make 10-1 dilution. From 10-1 dilution, 1ml of the mixture was transferred to the second tube and also swirled gently to make 10-2 dilution. These procedures were taken for the successive dilutions in a similar way to give 10-3, 10-4, 10-5, and 10-6 dilutions with the aid of separate sterile syringes. The diluent was inoculated on culture plates of nutrient agar, using the pour plate method. The plates were incubated in an inverted position for 24-48 hours at 37˚C. Colonies were counted and colony-forming unit per ml calculated. Colonies were subcultured to obtain a pure culture and were stored in 20% glycerol at 4 ˚C for further test.
2.4. Isolation on Selective Media
Isolation on two selective media, Salmonella/Shigella agar (SSA) and eosin methylene blue agar (EMBA) was performed for Salmonella, Shigella and E. coli isolation. The media were prepared according to the manufacturer’s instructions. Briefly, 1ml 10-1 to 10-3 dilutions were poured into a different petri dish containing SSA. The petri dish was swirled to ensure proper mixing of inoculum in the agar. This procedure was repeated for isolation on EMBA. All plates were then incubated for 24 to 48h at 37˚ C. After incubation, the colonies were subcultured on fresh plates to obtain pure cultures used for further identification. All experiments were performed in duplicates.
2.5. Morphological and Biochemical Identification of Isolates
Gram staining was done to differentiate organisms based on the structure of their cell wall [3]. Furthermore, bacterial isolates were identified based on their cultural, morphological and biochemical characteristics according to Bergey’s Manual of Determinative Bacteriology [8, 9]. Biochemical characteristics were determined using a conventional biochemical test such as catalase, coagulase, indole, and citrate tests [10].
2.5.1. Catalase Test
Few drops of 3% hydrogen peroxide were poured into a test tube, and a colony of the test organism was picked and suspended in the test tube [11]. Production of the bubble indicates a positive catalase test. Catalase test is used to differentiate the bacteria that produce the enzyme catalase, such as Staphylococci from non-catalase producing bacteria such as Streptococci.
2.5.2. Coagulase Test
A drop of distilled water was placed on each end of a clean grease-free slide, and a colony of the test organism was emulsified in each of the drops to make two thick suspensions and mixed gently. Plasma (from sheep blood) was added to one of the suspensions [12]. The clumping of the organism after 10 seconds indicates a positive result. Coagulase test is used to identify Staphylococcus aureus, which produces the enzyme coagulase.
2.5.3. Indole Test
It is used to determine the ability of an organism to split indole from the amino acid tryptophan using the enzyme tryptophanase. Tryptophan broth was inoculated with test organism and incubated for 24 h. Drops of Kovac's reagent were added to the broth. The formation of a red ring at the surface of the broth signifies a positive result [13]. Enterobacteriaceae produces indole hence the positive result with Kovac's reagent (Loba Chemie, India).
2.5.4. Citrate Utilisation Test
This test is based on the ability of the organism to use citrate as its source of carbon. Simmon citrate agar (Oxoid, Hampshire, England) medium was prepared as slants. A sterile wire loop was used to inoculate the isolates into the slants and incubated at 37˚C for 24 h after which it was examined for colour change [14] A change in colour of the medium to bright blue colour gives a positive citrate utilisation test while green colouration is considered negative.
2.6. Antibiotic Sensitivity Testing
Isolated bacteria from the fish pond in Ile- Ife were tested for antimicrobial susceptibility by the disc diffusion method. The panel of antibiotics (Abtek Biologicals Ltd, UK) used for testing were ceftazidime (CAZ) 30µg, cefuroxime (CRX) 30µg, gentamicin (GEN) 10µg, cefixime (CXM) 5µg, ofloxacin (OFL) 5µg, augmentin (AUG) 30µg, nitrofurantoin (NIT) 300µg, ciprofloxacin (CPR) 5µg. The susceptibility of the isolates was tested using the Kirby-Bauer disc diffusion method on Mueller Hinton agar (BIOTEC, Ltd), as described by Iwu et al. [15]. Briefly, 4-5 colonies of pure bacterial isolates were emulsified into a test tube containing 2ml of sterile normal saline (0.85% NaCl) using a sterilised flexible loop and homogenised to give turbidity that is equivalent to 0.5 McFarland standards (equivalent to 1.5 × 108 cells). A sterile cotton swab stick was dipped into the suspension, drained to remove excess culture and then streaked on the entire surface of the Mueller Hinton agar (MHA) plate. The inoculated plates were left on the bench for 3-5minutes to dry. The antibiotics discs were properly placed aseptically on the surface of the inoculated plates using a sterile forceps and gently to ensure even contact with the medium. After that, the plates were left on the bench for about 5minutes to allow the antibiotic’s diffuse into the medium and incubated at 37°C for 18 to 24 hours. The zone of inhibition around the disc was measured, and the result was interpreted as resistance or susceptible based on the interpretative standard according to the Clinical and Laboratory Standards Institute guidelines [16].
3. RESULTS
3.1. Physical Parameters and pH
The temperature value of the pond water sample from all sampling stations throughout the study period values ranged from 25˚C to 28˚C. The water sample from E1 and T1 had the highest value of 28˚C, respectively while C1 and E1 had the lowest value of 25˚C (Table 2). The water sample of T2 recorded the highest pH value of 9.0, while C1 has the lowest pH value of 7.1.
3.2. Total Heterotrophic and Total Coliform Bacteria Population
The total bacteria count of the water sample from sampling stations C1 to T2 ranged from 1.9×104 CFU/ml to 5.4×104 CFU/ml (Table 2). The mean values of the bacteria count for the six sampling stations from C1 to T2 were 4.9×104, 4.9×104, 5.4×104, 2.5×104, 2.2×104, and 1.9×104 CFU/ml, respectively. The bacterial count of the water sample from T2 recorded the lowest with the value of 1.9 ×104 CFU/ml, while the highest bacterial count was recorded from E1 with the value of 5.4×104 CFU/ml. The total coliform of the water sample of tank pond 2 has the lowest coliform count of 1.4 x 104 CFU/ml, while concrete pond 1 and earthen pond 1 has the highest coliform count value of 5.4 x 104 CFU/ml (Table 2).
3.3. Bacteria Types Detected in Water Samples
The cultural, morphological and biochemical characteristics of dominant bacteria isolated from the various sampling stations during the study are shown in (Tables 3-5). A total of 40 isolates of which 5 species were Gram positive bacteria representing two genera, and 35 species of Gram negative bacteria representing four genera were isolated from the water samples of the six sampling points. Isolates C101 to C102 obtained from C1 were identified as Escherichia coli, and Klebsiella sp., respectively. Isolate C201 from C2 was identified as Salmonella sp. Isolates E101 and E102 from E1 were identified as Klebsiella sp. Isolates E201 to E210, isolates T101 to T114 and isolates T201 to T211 were isolated from E2, T1 and T2 respectively, and the dominant bacteria found in these ponds were identified as Staphylococcus aureus, Enterococcus sp., Enterobacter sp., Klebsiella sp., Salmonella sp. and Escherichia coli.
| Pond type | Temperature °C | pH | Total heterotrophic Count (CFU/ml) | Total Coliform Count (CFU/ml) | |
|---|---|---|---|---|---|
| C1 | 25 | 7.1 | 4.9×104 | 5.4×104 | |
| C2 | 27 | 7.8 | 4.9×104 | 4.8×104 | |
| E1 | 25 | 8.5 | 5.4×104 | 5.4×104 | |
| E2 | 28 | 8.8 | 2.5×104 | 1.5×104 | |
| T1 | 28 | 8.1 | 2.2×104 | 1.9×104 | |
| T2 | 27 | 9.0 | 1.9×104 | 1.4×104 | |
| Isolate Code |
Cultural Characteristics on Agar | Cell Shape | Gram Reaction | Catalase | Citrate | Indole | Coagulase | Suspected Organism |
|---|---|---|---|---|---|---|---|---|
| C101 | Pink, round, entire flat on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| C102 | Greenish metallic sheen, irregular, entire, flat on EMB agar | R | - | N.A | - | + | - | E. coli |
| C201 | Black, round, entire, flat on SSA | R | - | N.A | + | - | - | Salmonella sp. |
| E101 | Pink, round, entire, convex on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| E102 | Pink, round, entire, flat on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| E201 | Yellow, round, entire, flat on NAG | C | + | + | + | - | + | S. aureus |
| E202 | Cream, irregular, entire, raised on NAG | C | - | N.A | + | - | - | Enterococcus sp. |
| E203 | Blue. round, entire, flat on EMB agar | R | - | N.A | + | - | - | Enterobacter sp. |
| E204 | Purple, round, entire, flat on EMB agar | R | - | N.A | + | - | - | Enterobacter sp. |
| E205 | Pink, round, entire flat on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| E206 | Pink, round, entire, raised on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| E207 | Pink, round, entire, flat SSA | R | - | N.A | - | + | - | E. coli |
| E208 | Pink, round, entire flat on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| E209 | Black, round, entire, flat on SSA | R | - | N.A | + | - | - | Salmonella sp. |
| E210 | Purple, round, entire, raised on EMB agar | R | - | N.A | + | - | - | Enterobacter sp. |
| Isolate Code |
Cultural Characteristics on Agar | Cell Shape | Gram Reaction | Catalase | Citrate | Indole | Coagulase | Suspected Organism |
|---|---|---|---|---|---|---|---|---|
| T101 | Cream, round entire flat on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| T102 | Cream round entire raised on NAG | C | + | + | + | - | - | Staphylococcus sp. |
| T103 | Pink, irregular, entire, flat on SSA | R | - | N.A | - | + | - | E. coli |
| T104 | Black, round, entire, flat on SSA | R | - | N.A | + | - | - | Salmonella sp. |
| T105 | Pink, irregular, entire, flat on SSA | R | - | N.A | - | + | - | E. coli |
| T106 | Black, round, entire, flat on SSA | R | - | N.A | + | - | - | Salmonella sp. |
| T107 | Greenish metallic sheen, irregular, entire, flat on EMB agar | R | - | N.A | - | + | - | E. coli |
| T108 | Blue, round, entire, flat on EMB agar | R | - | N.A | + | - | - | Enterobacter sp. |
| T109 | Purple, irregular, entire, flat on EMB agar | R | - | N.A | + | - | - | Enterobacter sp. |
| T110 | Black, round, entire, flat on SSA | R | - | N.A | + | - | - | Salmonella sp. |
| T111 | Pink, round, entire, raised on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| T112 | Purple, irregular, entire, flat on EMB agar | R | - | N.A | + | - | - | Enterobacter sp. |
| T113 | Greenish metallic sheen, irregular, entire, flat on EMB agar | R | - | N.A | - | + | - | E. coli |
| T114 | Pink, round, entire, raised on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| T201 | Yellow, round, entire, flat on NA | C | + | +ve | + | - | + | S. aureus |
| T202 | Yellow, round, entire, flat on NA | C | + | +ve | + | - | + | S. aureus |
| T203 | Pink, irregular, entire, flat on SSA | R | - | N.A | _ | + | - | E. coli |
| T204 | Black, round, entire, flat on SSA | R | - | N.A | + | - | - | Salmonella sp. |
| T205 | Pink, round, entire, raised on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| T206 | Blue, round, entire, flat on EMB agar | R | - | N.A | + | - | - | Enterobacter sp. |
| Isolate Code |
Cultural Characteristics on Agar | Cell Shape | Gram Reaction |
Catalase | Citrate | Indole | Coagulase | Suspected Organism |
|---|---|---|---|---|---|---|---|---|
| T207 | Black, round, entire, flat on SSA | R | - | N.A | + | - | - | Salmonella sp. |
| T208 | Pink, round, entire, raised on EMB agar | R | - | N.A | + | - | - | Klebsiella sp. |
| T209 | Greenish metallic sheen, irregular, entire, flat on EMB agar | R | - | N.A | - | + | - | E. coli |
| T210 | Purple, round, entire, raised on EMB agar | R | - | N.A | + | - | - | Enterobacter sp. |
| T211 | Greenish metallic sheen, irregular, entire, flat on EMB agar | R | - | N.A | - | + | - | E. coli |
3.4. Antibiotics susceptibility testing
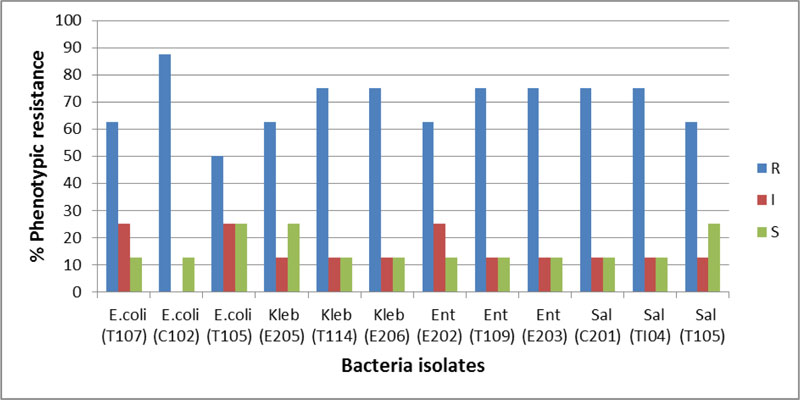
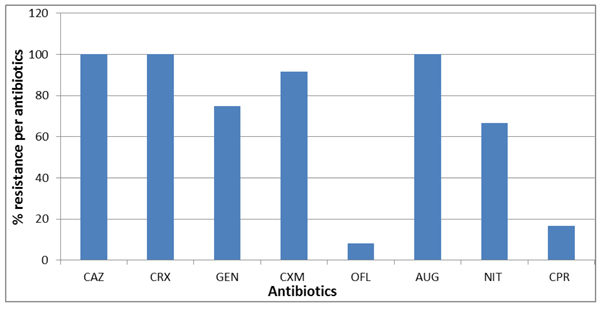
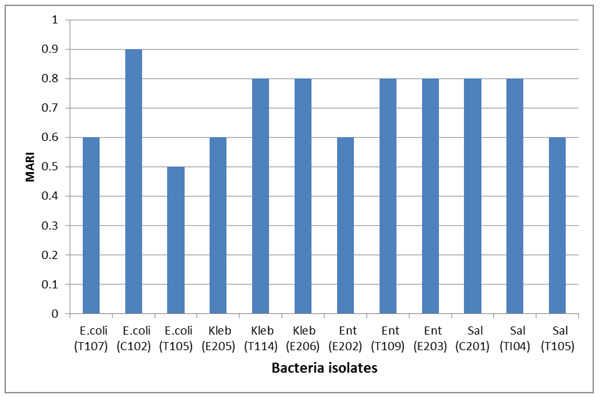
All isolates were 100% resistant to ceftazidime, cefuroxime and augmentin. More resistance to cefixime (80%) and gentamicin (73.3%) and nitrofurantoin (66.7%) was recorded (Figs. 2 and 3). However, only 16.6% and 8.3% of the isolates were resistant to ciprofloxacin and ofloxacin, respectively. The multiple antimicrobial resistance index (MARI) ranged from 0.5 to 0.9 (Table 6).
Kleb: Klebsiella sp., Ent: Enterobacter sp., Sal: Salmonella sp., Staph: Staphylococcus aureus
| Isolate (code) | Antibiotic susceptibility | Susceptibility % | MARI | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CRX | GEN | CXM | OFL | AUG | NIT | CPR | R | I | S | ||
| E.coli (T107) | R | R | R | R | I | R | S | I | 62.5 | 25 | 12.5 | 0.6 |
| E.coli (C102) | R | R | R | R | R | R | S | R | 87.5 | 0 | 12.5 | 0.9 |
| E.coli (T105) | R | R | S | R | S | R | I | I | 50 | 25 | 25 | 0.5 |
| Kleb (E205) | R | R | R | R | S | R | S | I | 62.5 | 12.5 | 25 | 0.6 |
| Kleb (T114) | R | R | R | I | S | R | R | R | 75 | 12.5 | 12.5 | 0.8 |
| Kleb (E206) | R | R | R | R | S | R | R | I | 75 | 12.5 | 12.5 | 0.8 |
| Ent (E202) | R | R | I | R | S | R | R | I | 62.5 | 25 | 12.5 | 0.6 |
| Ent (T109) | R | R | R | R | S | R | R | I | 75 | 12.5 | 12.5 | 0.8 |
| Ent (E203) | R | R | R | R | S | R | R | I | 75 | 12.5 | 12.5 | 0.8 |
| Sal (C201) | R | R | R | R | S | R | R | I | 75 | 12.5 | 12.5 | 0.8 |
| Sal (TI04) | R | R | R | R | S | R | R | I | 75 | 12.5 | 12.5 | 0.8 |
| Sal (T105) | R | R | S | R | S | R | R | I | 62.5 | 12.5 | 25 | 0.6 |
| Resistance % | 100 | 100 | 75 | 91.6 | 8.3 | 100 | 66.6 | 16.6 | ||||
| Mean ±SD | 69.7±9.9 | 14.5±7.2 | 15.6±5.6 | 0.7±0.1 | ||||||||
| T-value | 24.3 | 7 | 9.6 | 19.6 | ||||||||
R: Resistance, I: Intermediate, S: Susceptible, Kleb: Klebsiella sp., Ent: Enterobacter sp., Sal: Salmonella sp., Staph: Staphylococcus aureus
CAZ: Ceftazidime (30 μg), CRX: Cefuroxime (30 μg), GEN: Gentamicin (10 μg), CXM: Cefixime (5 μg), OFL: Ofloxacin (5 μg), AUG: Augmentin (30 μg), NIT: Nitrofurantoin (300 μg) and CPR: Ciprofloxacin (5 μg)
4. DISCUSSION
The pH (7.1-9.0) and temperature (25-28˚C) recorded in all the ponds were within the optimum range necessary for African fish aquaculture [17, 18]. pH water quality parameter is vital in fish aquaculture as it affects the toxicity of other compounds to fish, including chlorine and ammonia [19]. Temperature is often referred to as the principal factor among environmental factors affecting aquatic life [20], as it affects all biochemical processes with direct consequences, including food requirement and conversion efficiency, and fish growth. Optimum pH and temperature directly stabilise the physicochemical parameters of pond water, enhancing fish health and productivity and the maintenance of a proper balance of the microbial ecology in pond water [21]. A lower pH range (pH 6.75-7.10) than that of this study, however, was reported in water samples collected from concrete and earthen fish ponds in Edo State, Nigeria [22]. Shoko et al. [23] reported a pH range of 6.5-7.9 in earthen pond of mono- and poly-culture of African sharp tooth catfish (Clarias gariepinus, Burchell, 1822). Makori et al. [21] reported temperature and pH range of 24-26 °C, and pH 6.1–8.3, respectively. A study conducted in Kenya, reported a temperature range from 20.04˚C to 32.63˚C [24], elsewhere, the temperature was lower than in the present study [7]. Rapid and fluctuating changes in pH and temperature have been reported to cause extreme stress in the fish.
In the present study, more Gram-negative bacteria were isolated than Gram-positive from the various fish ponds water. However, the total bacterial count could be an underestimation due to viable but non-culturable (VBNC) state of some microbial population. The bacteria isolated were E. coli, Salmonella sp., Klebsiella spp., Enterobacter spp., Staphylococcus spp., and Enterococcus spp. The bacterial load observed in water could be a result of the effect of anthropogenic activities in the location where the ponds are situated. Moreover, the use of chicken manure as feed by some fish farmers could serve as easy biodegradable organic matter, which may increase the total bacterial count [25, 26]. The coliforms isolated were an indication of the contamination of the pond water with faecal materials. Some reports have indicated high incidence of antibiotic resistant bacteria in integrated fish farming; a practice that combines aquaculture with other livestock farming operations and use of their manure for fish feed [27-29]. Faecal contamination of pond water may result in the introduction of pathogenic organisms into the pond. Such pathogens have been shown to lead to fish diseases or foodborne disease [2, 3, 30].
Furthermore, the fertilisation of the ponds with animal manure poses the risk of introduction of potentially pathogenic and resistant bacteria into the pond. The resistant bacteria could infect fishes, humans and contaminate the environment. Animal faeces have been shown to harbour a plethora of potentially dangerous bacteria, some of which are resistant to antibiotics of choice used in the treatment of human and animal disease [15, 31]. Resistant bacteria are capable of transferring and conferring resistance to commensal bacteria through horizontal gene transfer using mobile genetic elements (MGE) such as conjugative plasmids, transposons, and phages [32]. The Enterococci species, in particular, are among the major etiological agents of nosocomial infection, and are prone to acquisition of resistance determinants [33].
The different groups of bacteria isolated from these fish ponds are in concordance with studies conducted elsewhere [13, 34-36]. Contaminated feed added to the ponds introduces allochthonous bacteria, which subsequently become the principal source of bacteria contaminating pond water and affecting fish health. The presence of pathogenic microorganisms, especially Enterobacter spp., E. coli, Salmonella and Staphylococcus spp. can lead to the transmission of water-borne diseases such as Typhoid fever, Cholera, food poisoning and gastroenteritis in humans. Often the main route of disease transmission is the consumption of improperly cooked fish cultivated in these ponds [30, 36].
The antibiotics susceptibility test (AST) of the studied coliforms shows that 100% of the isolates were 100% resistant to Ceftazidime, Cefuroxime and Augmentin; 73.3% were resistant to Gentamicin. Up to 80% of the isolates were resistant to Cefixime, 6.7% were resistant to Ofloxacin. Also, 66.7% were resistant to Nitrofurantoin. The AST result corresponds to the findings in Singapore where 100.0%, 97.0%, and 97.0% of isolates were resistant to cefazolin, ceftazidime, and cefepime, respectively [37]. On the contrary, Onuoha [38] reported that resistance for nitrofurantoin, clarithomycin and ampicillin was widespread in bacteria from aquaculture sources of south eastern Nigeria. A similar result was obtained with V. parahaemolyticus, isolates in Malaysia, where high resistance to ampicillin, amikacin, kanamycin, cefotaxime, and ceftazidime was reported [39]. The highest (85%) and lowest (1.1%) resistance towards streptomycin and gentamicin, respectively, was reported in bacteria recovered from aquaculture in Malaysian Borneo [40]. In another study, a relatively low prevalence of antibiotic resistance was detected in bacteria from a marine fish cage-culture area of Guangdong China. A high proportion of oxytetracycline-resistant bacteria, however, was recorded in the study [41]. Another study reported the isolation of several fish pathogenic bacteria including Plesiomonas shigelloides, Aeromonas sobria, Vibrio cholera and Klebsiella spp., which all expressed high levels of resistance to penicillin, oxacillin and ampicillin [42].
The 100% resistance to augmentin found in this study, did not come as a surprise given that augmentin is listed among the top 20 antibiotics that contribute 61% towards the total systemic antibacterial market [43]. Nonetheless, a report on an outbreak of foodborne disease among school teachers at Rob Ferreira High School in White River, Mpumalanga, South Africa, found no resistance among S. enterica serotype Virchow [44]. However, a 14% resistance to augmentin was reported in Salmonella typhi antibiotic sensitivity pattern in Dubai, United Arab Emirates [45]. In addition to AMR being a global public health problem, Nigeria also faces a precarious situation of poor antibiotic prescription monitoring and prescription-only medicines (POM), including antimicrobials that are routinely sold Over-The-Counter (OTC) in pharmacies and by patent proprietary medicines vendors (PPMVs). Easy access to critical antibiotics of last resort by human patients increases its imprudent use, further worsening the problem of AMR. Currently, there are no available studies outlining the full burden of AMR and its health and economic impact on Nigerians [46]. Hence, the implication of the finding of this study could be far-reaching and could be used as a baseline study on AMR in the environment.
In this study, the multiple antibiotic resistance indexes for all isolated bacteria ranged from 0.5 to 0.9, indicating a high-risk environment (Fig. 4). A study with a similar finding was conducted in southeastern Nigeria on the prevalence of antibiotic-resistant diarrhoeagenic bacterial species in Surface Waters. The result of the Nigerian research showed that Salmonella spp. had the highest MARI value of 0.75, whereas the least MARI value of 0.44 was obtained for Staphylococcus spp [47]. Also, in Benin City Nigeria, multiple antibiotic resistance index mean value of 0.365 for Vibrio isolates obtained from four different fish pond facilities was reported [48].
Antimicrobial resistance is a global public health problem sustained by multifactorial antibiotic usage. The spread of antibiotic resistance in the marine environment is poorly understood [49]. Less attention has been given to the aquatic ecosystem in the transmission of AMR bacteria to humans. This has created inadequate information on the antimicrobial drug susceptibility of the aquatic environment, which is vital in the epidemiology of AMR [42]. However, antibiotic resistance genes (ARGs) in environmental reservoirs and the transferability of these genes through MGE would likely account for the growing prevalence of ARGs in the aquaculture settings. For instance, Huang et al. [50] reported much higher total concentrations of detected ARGs of about 1.6–4.0 times in culture pond water than the control, as well as the presence of motility genes. Furthermore, global usage of antimicrobial agents to control infections in humans and animals is a common practice. In these circumstances, antimicrobial agents used in marine and aquaculture systems not only escalates the pathogenic bacterial resistance to antibiotics but also increases the likelihood of resistance genes transfer to common commensal bacteria, thus promoting resistance to multiple antibiotics [51].
Kleb: Klebsiella sp., Ent:Enterobacter sp., Sal: Salmonella sp., Staph: Staphylococcus aureus
CONCLUSION
The concrete pond (C1 and C2), and earthen (E1) pond had the highest load of heterotrophic and coliform bacteria. The water quality parameters (temperature and pH) and the type of bacteria detected in all ponds did not differ significantly. The study also revealed that all the ponds were contaminated with potentially pathogenic bacteria that could affect fish health and fish product. These organisms could lower fish yield, cause diseases and economic loss, and equally endanger public health, particularly if the fish harvested from the ponds are not properly cooked before consumption. Discharging of contaminated pond water into the environment could further enhance the transfer of resistant bacteria to the environment. Hence, the regular monitoring of pond water for microbial contamination is necessary to maintain fish and public health. Good agricultural practices, such as the use of good quality water, regular draining of pond water after a specific period, and closure of ponds to the public, will aid in preventing pond water contamination. Moreover, 90% of the world's aquaculture production is carried out in developing countries. They mostly lack regulations and enforcement on the use of antibiotics, thereby necessitating the development and enforcement of such regulatory systems [52]. The use of modern techniques such as polymerase chain reaction and matrix-assisted laser desorption/ionisation (MALDI) used with time-of-flight mass spectrometer (TOF) to quantify and characterise bacteria from pond water is necessary for accurate and timely identification.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not Applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The authors confirm that the data supporting the findings of this study are available within the article.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Authors are grateful to the owners of the aquaculture facility survey in the study and also to the support staff of the Department of Biological Sciences, College of Natural and Applied Sciences, Oduduwa University Ipetumodu, Ile Ife, Osun State.


