All published articles of this journal are available on ScienceDirect.
In Vitro Antagonistic Activity of Diverse Bacillus Species Against Fusarium culmorum and F. solani Pathogens
Abstract
Background:
Fusarium culmorum and Fusarium solani are economically important fungal pathogens of many plant species causing significant yield losses worldwide. Frequent uses of fungicides are hazardous to humans and the environment. Therefore, in vitro antagonistic activity of diverse Bacillus species isolates with biological potential activity to control these both pathogens should be investigated.
Objective:
The objectives were to isolate and identify the Bacillus spp., which are potential controls of F. culmorum and F. solani, and to characterize molecularly, at the species level, those isolates that have potential as biocontroller of the pathogens.
Methods:
The in vitro antagonistic potential of 40 Bacillus isolates against F. culmorum and F. solani was evaluated on the basis of fungal growth inhibition on nutrient broth culture. The colony morphology and the 16S rRNA gene sequencing of Bacillus spp. were used to identify the isolates.
Results:
Bacillus sp. isolates were identified as B. atrophaeus, B. subtilis, Paenibacillus polymxa, B. amyloliquefaciens, B. simplex and B. tequilensis. They had significant (P<0.05) antagonistic activities against F. culmorum and F. solani isolates as compared to the untreated control. The antagonistic effects varied depending on the Fusarium sp. The bacterial B. subtilis isolates SY116C and SY SY118C provided the most noteworthy result as both strongly inhibited mycelial growth of F. solani by 97.2%, while the B. tequilensis isolate SY145D was the most effective in the formation of inhibition zones against F. culmorum by 75%.
Conclusion:
It is apparent that Bacillus sp. isolates play an important role in the inhibition of growth of F. culmorum and F. solani, and that the B. subtilis isolates SY116C and SY118C had the highest biological potential activity against these fungi. These antagonistic effects may be important contributors as a biocontrol approach that could be employed as a part of integrated soil pathogen management system.
1. INTRODUCTION
Fusarium culmorum and Fusarium solani are important species that cause significant yield losses in many plant species [1, 2]. Fusarium culmorum is a soil-borne fungus distributed in cooler temperate regions, and the causal agent of many important diseases in cereals [3, 4]. F. solani is known to infect many plant species, causing plant decline, wilting, and large necrotic spots on tap roots [5, 6].
Fungicides are commonly used to control these diseases, but frequent uses of these chemicals are hazardous to humans and the environment. Therefore, the management of the soil-borne pathogens has become one of the major concerns in agriculture and focused on searching and selecting antagonist microorganisms on diverse soil pathogens [7, 8]. However, difficulties in controlling Fusarium sp. have stimulated renewed interest in the application of biological control agents, which has provided an effective and environmentally friendly means to control plant diseases. Among the most widely used microbes for biocontrol agents are members of the genus Bacillus, that offer advantages over other microorganisms, tolerance to fluctuating pH, temperature and osmotic conditions [9, 10]. Furthermore, Bacillus spp. are able to colonize root surfaces, promoting plant growth and causing mycelium lysis of several fungal agents [11, 12].
However, soil-borne fungal pathogens such Fusarium sp. have been shown varying metabolic responses, growth patterns and reproductive strategies in response to varying soil microorganisms, therefore, measuring the in vitro growth rate of fungi was considered as a simple and reliable method for evaluating the effect of an environmental variable on the growth of fungi, although this does not take into account changes in mycelia density [13].
On the other hand, the 16S ribosomal RNA (16S rRNA) gene has been widely used for the taxonomic classification of bacteria by the detection of sequence differences in the hypervariable regions of the 16S rRNA gene which is present in all bacteria [14, 15].
During a polyphasic experiment, more than 525 bacilli were isolated from different regions of Syria. In the present study, forty of them were taken into the 16S rRNA gene sequence analyses.
The objectives of this current work were to isolate and identify the bacteria which have the potential to control Fusarium spp. (F. culmorum and F. solani) and molecular characterization of species that have the potential for biocontrol of the pathogen.
2. MATERIALS AND METHODS
2.1. Isolates of Bacillus sp.
The soil samples were collected from different regions distributed widely from south to the north-west between 33.40°N and 37.17°N and 35.40°E and 42.30°E in Syria. They were taken from 2-3 cm depth of field and were carried to laboratory in sterile polythene bags. They were shaken in 9 ml sterile water for 3 min at 160 rpm. Serial dilution was made from 10-3 to 10-7 [16] and then 0.2 ml of each dilution was spread onto Nutrient Agar (NA) medium and incubated overnight, the colonies of prospective Bacillus sp. were identified according to Wulff et al. [17], and 40 isolates were selected for this study (Table 1). Six Bacillus species, namely, B. atrophaeus, B. subtilis, P. polymxa, B. amyloliquefaciens, B. simplex and B. tequilensis, were selected for the further in vitro study. A pure culture of each Bacillus sp. isolate was first grown on NA and incubated for 24h at 30°C.
2.2. Fungal Isolates
Fusarium culmorum and F. solani were isolated from infected wheat plants growing in different locations of Syria. The infected wheat stems were cut into small pieces of 1-1.5 cm, surfaces were sterilized with 5% sodium hypochlorite for 5 min, washed in sterile distilled water twice and cultured on PDA (PDA, DIFCO, Detroit, MI. USA) medium amended with 13 mg/l kanamycin sulphate added after autoclaving and incubated for 10 days, at 23 ±1ºC in the dark to allow mycelial growth and sporulation. Species identification was based on the morphological characteristics of single spored isolates as described by Nelson et al. [18, 19]. According to a study [20], the two virulent monosporic isolates of F. culmorum and F. solani were selected for this study. The cultures were maintained on silica gel at 4 ºC until needed.
| Bacillus species | Number of isolates | Morphology |
|---|---|---|
| B. atrophaeus | 3 | Brown-black, opaque, smooth, circular |
| B. amyloliquefaciens | 10 | Creamy white with irregular margins |
| P. polymyxa | 2 | Milky white, thin often with amoeboid spreading |
| B. subtilis | 20 | Fuzzy white, opaque, rough, with jagged edges |
| B. simplex | 1 | Cream, gloss, with irregular margins slightly raised |
| B. tequilensis | 4 | Yellowish, opaque, smooth, circular |
2.3. In vitro Activity of Bacillus spp. Isolates against Fusarium spp.
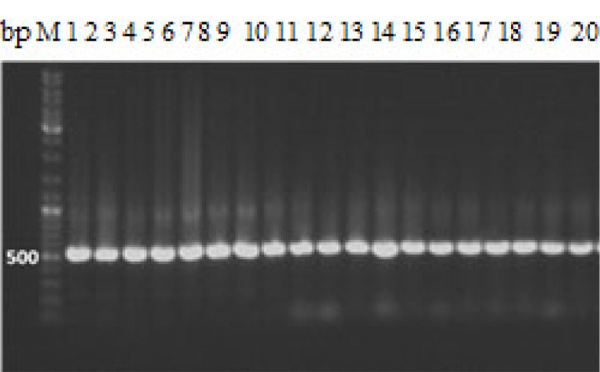
Bacterial colonies of different size, color and morphology were streaked individually a few times until single colonies of a single type were observed on the NA plates. Then 5 mm diameter disc of each F. culmorum and F. solani was cut from of an actively growing culture by a sterile cork borer and placed onto the center of above NA plates. Where mycelia disc on Nutrient Agar (NA) medium without bacteria was maintained as control. Every elementary treatment was repeated five times. The mean diameter of pathogen colonies was measured after 4 days of incubation at 25°C and any morphological alteration of colonies, in comparison to the untreated control, was also noted. Damage caused by the bacterium to the fungal mycelium, removed from the confrontation zone of both microorganisms (pathogen and antagonist), was observed under a light microscope, in comparison to untreated controls. The percentage of inhibition of fungal growth was calculated by the following formula proposed by Rabindran and Vidyasekaran [21].
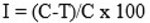 |
Where; I = Percent inhibition, C = Radial growth of the pathogen in control, and T = Radial growth of the pathogen in treatment.
2.4. 16S rRNA Gene Sequencing
Selected bacterial 16S rRNA was amplified in full length by PCR using two pairs of primers, BacF (5’-GTGCCTAATACATGCAAGTC-3’) and BacR (5'- CTTTACGCCCAATAATTCC-3’) [12]. The PCR reaction mix (50 μl) contained 2 μl (50-100 ng) of extracted genomic DNA, 1x reaction buffer (TrisKCl-MgCl2), 2 mM MgCl2, 0.2 mM dNTP, 1 μM of each primer, and Taq polymerase (5U/μl, Fermentas). PCR amplification condition was achieved using the following parameters: An initial denaturation step at 95°C for 5min followed by a second denaturation step at 95°C for 1min, annealing for 1min at 54°C, an extension at 72°C for 90s, and a final extension step of 72°C for 10min. A total of 30 serial cycles of amplification reaction were performed. PCR products were separated on a 1.5% agarose gel and visualized using UV light (302 nm) after staining with ethidium bromide. Prior to sequencing, PCR products were purified with QIAgen gel extraction kit (28704) according to the manufacturer´s recommendations. Sequencing was carried out on a Genetic Analyzer (ABI 310, Perklin-elmer, Applied Biosystems, USA). The 16S rRNA sequences were compared with the known sequences using the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih. gov).
2.5. Statistical Analyses
All experiments were performed in triplicate with ten Petri dishes per replicate, for each bacterium-fungus in vitro evaluation, using a completely randomized design. An F-test was used to determine if the two runs of each experiment were homogeneous and if the data could be pooled. The homogeneity of variance test indicated that the data from both runs of each experiment could be pooled, and thus all further analyses were conducted on pooled data. Data were analyzed using analysis of variance (ANOVA) and means were separated by Tukey's test (P≤ 0.05).
3. RESULTS AND DISCUSSION
In this present work, the antagonistic potential of the Bacillus sp. isolates was concluded and validated by restriction of the F. culmorum and F. solani pathogens growth and showed zone of inhibition towards the antagonist as shown in photo-plate of NA culture plate assay compared with the control (Fig. 1).
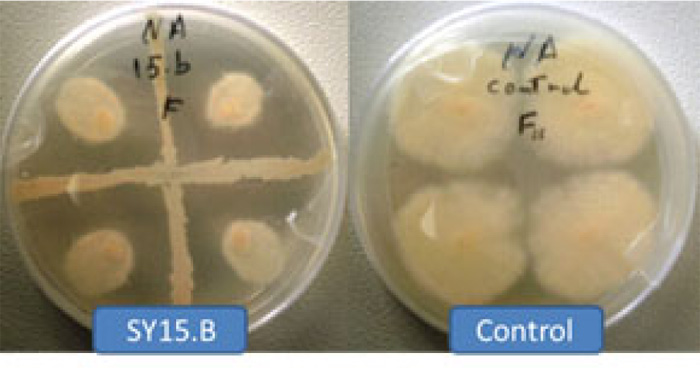
On the other hand, PCR amplification with specific primers Bac yielded single DNA fragments of ~ 545 bp, present in all Bacillus sp. isolates (Fig. 2). On the basis of 16S rRNA gene sequencing, Bacillus sp. isolates are identified as B. atrophaeus, B. subtilis, P. polymxa, B. amyloliquefaciens, B. simplex and B. tequilensis as their 16S rRNA gene sequences displayed similarities ≤ 98% to their closely related type strains (Table 2). Species belonging to the Paenibacillus genus were previously re-classified under the genus Bacillus, based on morphological characteristics. However, the PCR probe tests suggested that a group of eleven species should be considered a new genus, Paenibacillus, of which P. polymyxa is the type strain [22]. The nucleotide sequences were deposited in GenBank under accession numbers MT159352 to MT159391 (Table 3).
| Bacillus species | Microorganisms (NCBI) | 16S rRNA gene sequence similarity (%) |
|---|---|---|
| B. atrophaeus | B. atrophaeus, ATCC 49337 | 96% |
| B. simplex | B. simplex JP44SK12 (JX144702) | 97% |
| B. subtilis | B. subtilis subsp. Spizizenii JP44SK23 (JX144713) | 99% |
| P. polymxa | B. paenibacillus macqariensis subsp. Defensor (AB360546) | 98% |
| B. amyloliquefaciens | B. amyloliquefaciens (AF478077) | 99% |
| B. tequilensis | B. Bacillus tequilensis (KT760402) | 98% |
| No. | Isolates | Zone of Inhibition (%)* | Antifungal activity | GeneBank |
| accession number | ||||
| B. atroplaeus | ||||
| 1 | SY15B | 45h* | + | MT159352 |
| 2 | SY199A | 46h | + | MT159353 |
| 3 | SY63E | 56.7e | + | MT159354 |
| B. subtilis | ||||
| 4 | SY35A | 55ef | + | MT159355 |
| 5 | Sy41B | 53f | + | MT159356 |
| 6 | SY44A | 50g | + | MT159357 |
| 7 | SY60A | 47gh | + | MT159358 |
| 8 | SY73B | 38i | + | MT159359 |
| 9 | SY113C | 60d | ++ | MT159360 |
| 10 | SY116C | 58.3d | + | MT159361 |
| 11 | SY118C | 53.3f | + | MT159362 |
| 12 | SY124B | 45h | + | MT159363 |
| 13 | SY130D | 46h | + | MT159364 |
| 14 | SY132E | 60d | ++ | MT159365 |
| 15 | SY133 | 63c | ++ | MT159366 |
| 17 | SY132C | 63.3c | ++ | MT159367 |
| 19 | SY134D | 61d | ++ | MT159368 |
| 20 | SY135D | 57.7e | + | MT159369 |
| 21 | SY139D | 36.7i | + | MT159370 |
| 22 | SY151C | 48.3g | + | MT159371 |
| 23 | SY160C | 66.7cd | ++ | MT159372 |
| 24 | SY168C | 66.7cd | ++ | MT159373 |
| 25 | SY190E | 43.3hi | + | MT159374 |
| Paenibacillus polymyxa | ||||
| 24 | SY53C | 60d | ++ | MT159375 |
| 25 | SY55B | 70b | ++ | MT159376 |
| B. amyloliquefaciens | ||||
| 26 | SY82C | 46h | + | MT159377 |
| 27 | SY96C | 66.7cd | ++ | MT159378 |
| 28 | SY96E | 61.7d | ++ | MT159379 |
| 29 | SY123A | 53.3f | + | MT159380 |
| 30 | SY128B | 63c | ++ | MT159381 |
| 31 | SY134C | 56.7e | + | MT159382 |
| 32 | SY159D | 61.7c | ++ | MT159383 |
| 33 | SY177C | 53.3f | + | MT159384 |
| 34 | SY190D | 51.7fg | + | MT159385 |
| 35 | SY200D | 57.7e | + | MT159386 |
| B. tequilensis | ||||
| 36 | SY69A | 46h | + | MT159387 |
| 37 | SY145D | 75a | +++ | MT159388 |
| 38 | SY149C | 71.7ab | ++ | MT159389 |
| 39 | SY150D | 55ef | + | MT159390 |
| B. simplex | ||||
| 40 | SY198B | 10k | + | MT159391 |
| LSD | 0.05 |
The data showed that Bacillus sp. isolates had a significant (P<0.05) antagonistic activity against both F. culmorum and F. solani where the percentage of radial growth inhibition of the fungi colonies significantly decreased on NA medium as compared to the untreated controls (Tables 3 and 4). In addition, the mean colony diameter of Fusarium sp., noted after 4 days of incubation at 25°C, depends upon pathogens tested and treatments realized, which is in agreement with a previous study [23]. However, the antagonistic effects of the bacterial isolates varied depending on the Fusarium spp. B. subtilis isolates SY116C and SY SY118C inhibited the growth of F. solani by 97.2% while the B. tequilensis isolate SY145D was the most effective in the formation of inhibition zones against of F. culmorum by 75% (Tables 3 and 4). Bacillus subtilis isolates showed similar antagonism against Rhizoctonia solani, Helminthosporium spp., Alternaria spp. and Fusarium oxysporum [24], and B. tequilensis isolate SY145D showed similar antagonism with B. tequilensis GYLH001 that had a potential antagonism towards Magnaporthe oryzae of rice [25]
| No. | Isolates | Zone of Inhibition (%)* | Antifungal activity |
| B. atroplaeus | |||
| 1 | SY15B | 76.8d* | +++ |
| 2 | SY199A | 73.9de | ++ |
| 3 | SY63E | 57.8gh | + |
| B. subtilis | |||
| 4 | SY35A | 75.4de | +++ |
| 5 | Sy41B | 87c | +++ |
| 6 | SY44A | 76.8d | +++ |
| 7 | SY60A | 100a | +++ |
| 8 | SY73B | 65.2fg | ++ |
| 9 | SY113C | 87.1c | +++ |
| 10 | SY116C | 97.2a | +++ |
| 11 | SY118C | 97.2a | +++ |
| 12 | SY124B | 100a | +++ |
| 13 | SY130D | 81.2cd | +++ |
| 14 | SY132E | 78d | +++ |
| 15 | SY133 | 76.9d | +++ |
| 17 | SY132C | 76.4d | +++ |
| 19 | SY134D | 68f | ++ |
| 20 | SY135D | 77d | +++ |
| 21 | SY139D | 76.2d | +++ |
| 22 | SY151C | 50h | + |
| 23 | SY160C | 71.2e | ++ |
| 24 | SY168C | 88.4c | +++ |
| 25 | SY190E | 71.5e | ++ |
| Paenibacillus polymyxa | |||
| 24 | SY53C | 27i | + |
| 25 | SY55B | 33.6i | + |
| B. amyloliquefaciens | |||
| 26 | SY82C | 87c | +++ |
| 27 | SY96C | 88.4c | +++ |
| 28 | SY96E | 94.5b | +++ |
| 29 | SY123A | 84cd | +++ |
| 30 | SY128B | 70.2e | ++ |
| 31 | SY134C | 71.2e | ++ |
| 32 | SY159D | 62g | ++ |
| 33 | SY177C | 73de | ++ |
| 34 | SY190D | 69.3f | ++ |
| 35 | SY200D | 74.5de | ++ |
| B. tequilensis | |||
| 36 | SY69A | 73.9de | ++ |
| 37 | SY145D | 72.4e | ++ |
| 38 | SY149C | 85.3cd | +++ |
| 39 | SY150D | 75.1de | ++ |
| B. simplex | |||
| 40 | SY198B | 10k | + |
| LSD | 0.05 |
It is well known that B. subtilis strains produce a broad spectrum of antimicrobial compounds, including predominantly peptides as well as a couple of non-peptidic compounds such as polypeptides, an aminosugar, and a phospholipid [26], and their highly antifungal effects (97.2%) on both F. culmorum and F. solani in this study, which might be attributed to one or more antifungal compounds produced by this biocontrol agent. However, the observed mycelial growth inhibition and lysis formation among the colonies of the both Fusarium pathogens might be due to the effect of the bacterial diffusible inhibitory antibiosis substances, which could have suppressed and restricted the growth of the pathogen, which can be confirmed by the fact that most Bacillus spp. have an ability to produce several antibiotics such as bacillomycin, fengycin, mycosubtilin and zwittermicin, which effectively suppress the growth of pathogens under in vitro and/or in situ conditions [27-29]. This might explain the formation of inhibition zones between the bacterial and the F. culmorum and F. solani isolates shown in this study. Our results are similar to those reported in a previous study [29, 30], which showed high capacity of some strains of Bacillus sp. of the same species to inhibit the growth of several phytopathogic fungi; this effect was attributed to the production and secretion of antifungal compounds and antibiotics belonging to the family of iturins and subtilins, that act on the fungi's cell wall [30]. Hence the most likely explanation for the growth reduction of F. culmorum and F. solani by Bacillus sp. is that antifungal activity is increased by co-culturing of different bacterial species.
CONCLUSION
Collectively, this work illustrates that two B. subtilis isolates (SY116C and SY SY118C) provided the most noteworthy result as both strongly inhibited mycelial growth of F. solani by 97.2%, and that one B. tequilensis isolate (SY145D) had a potent antagonistic activity of F. culmorum by 75%. These B. subtilis and B. tequilensis isolates, as potential biocontrol agents, may provide an effective strategy to combat plant pathogens. Field studies should be undertaken to confirm the effectiveness of the isolates under natural field conditions as a component of integrated disease management.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This work was supported by the Atomic Energy Commission of Syria.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors wish to thank the Director General of AECS and the Head of the Molecular Biology and Biotechnology Department for their continuous support throughout this work. Thanks are also extended to Dr. H. Ammouneh for his assistance in achieving the experiments. We also thank Dr. A. Al-Daoude for critical reading of the manuscript.


