RESEARCH ARTICLE
Coat Protein 1 Gene of Indonesian Isolates of Rice Tungro Spherical Virus showed High Divergence with those from other South and South East Asian Regions
Raden Heru Praptana1, Adyatma Irawan Santosa2, *, Rahmini Rahmini1, Wasis Senoaji1, Nur Rosida1, Danarsi Diptaningsari1, Affandi Affandi3, Riska Riska3, Y. Andi Trisyono2, Sedyo Hartono2
Article Information
Identifiers and Pagination:
Year: 2023Volume: 17
E-location ID: e187433152307140
Publisher ID: e187433152307140
DOI: 10.2174/18743315-v17-230811-2023-31
Article History:
Received Date: 18/04/2022Revision Received Date: 02/06/2023
Acceptance Date: 30/06/2023
Electronic publication date: 04/09/2023
Collection year: 2023

open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background:
Tungro, an important viral disease in rice, is a co-infection of rice tungro bacilliform virus (RTBV) and rice tungro spherical virus (RTSV). Severe symptoms were reported in rice fields on two major islands in Indonesia: Jawa (Magelang District) and Sulawesi (Sidrap District).
Objective:
Sequences of global isolates currently registered in NCBI GenBank were examined with two new Indonesian isolates to understand the phylogeny of RTSV based on different genome regions as well as to spot the gap in the molecular data of Indonesian isolates.
Methods:
Transmission test of RTSV using the vector green leaf hopper was performed, and then the complete sequences of coat protein 1 (CP1) gene of Magelang and Sindrap districts were molecularly studied using MEGA X software and Recombination Detection Program.
Results:
The obtained complete sequence of coat protein 1 (CP1) gene of Magelang-7 (OQ426495) and Sindrap-9 (OQ426496) isolates were aligned with 39 isolates in NCBI GenBank. Sequences of other isolates in GenBank were also retrieved and then aligned according to six individual Open Reading Frames (ORFs) and ‘Full ORF’. Recombination analysis detected significant signals in the genomes of one partially and four fully sequenced isolates. Phylogenetic study also confirmed the frequent recombination among RTSV isolates clustered in two geographically separated groups: South East Asia and South Asia.
Conclusion:
Although the available data is still rudimentary, this study demonstrated that the CP1 gene of 21 isolates from different regions of Indonesia was highly identical among themself but divergent with isolates from other countries. Therefore, deep molecular comprehension on Indonesian isolates is necessary to manage the recurrent Tungro spread in the country.
1. INTRODUCTION
A viral disease of rice (Oryza sativa L.), commonly named Tungro, is widespread in Southeast and South Asian countries and responsible for considerably large losses. Typical symptoms include leaf chlorosis and growth inhibition leading to plant dwarf. The disease is a manifestation of a synergy of two viruses: rice tungro bacilliform virus (RTBV) and rice tungro spherical virus (RTSV) [1]. Although the exact function of the protein encoded by RTBV ORF-IV is not known, its interaction with RTSV coat protein 3 (CP3) is thought to restrain the host RNA silencing mechanism, thus the need for both viruses’ presence in more intense Tungro cases [2]. On the other hand, RTSV plays an important role in the green leaf hopper (Nephotettix virescens D.) transmission of the viral complex [3].
RTSV is classified in the genus Waikavirus within the family Secoviridae. Genome of the virus is a polyadenylated single-stranded RNA with a length of ± 12 kb, encoding a large polyprotein that can be split into three mature coated proteins (CP1, CP2, and CP3) and a hypothetical proteins, NTP-binding domain, 3C-like protease, and RNA-dependent RNA (RdRp) proteins [4]. The genome is encapsidated in isometric shaped particles [1].
Co-infection of the two viruses as the cause of Tungro in Indonesia has been long established [5], and yet the disease is still a perpetual constrain on rice production in the country [6-8]. In this study, the full sequence of CP1 gene of two Indonesian isolates from Java and Sulawesi Islands was observed. Furthermore, the sequences of global isolates currently registered in NCBI GenBank were also examined to understand the phylogeny of RTSV based on different genome regions as well as to spot the gap in the molecular data of Indonesian isolates that need to be addressed in order to develop advanced management of the virus.
2. MATERIALS AND METHODS
2.1. RTSV Transmission
RTSV transmission test using its vector N. virescens was performed as a response to reports of widespread tungro-like symptoms on rice cultivated in Panca Rijang Subdistrict, Sidenreng Rappang (Sidrap) District, Sulawesi Selatan Province and Muntilan Subdistrict, Magelang District, Jawa Tengah Province of Indonesia in 2011 (Fig. 1).
 |
Fig. (1). Rice plants on fields in (A). Magelang, Jawa Tengah, and (B). Sindrap, Sulawesi Selatan showed severe Tungro symptoms: leaf yellowing and stunting. |
The test procedures were similar to those of another study [9], with some modifications. Seeds of rice variant TN1 were sown on soil in pots and then grown for 10 days after germination. Each selected healthy seedling was then put in a small glass test tube filled with distilled water up to 1 cm and sealed with cotton. Ten tubes with rice seedlings were then brought to each sampling location. In each location, one small clump of rice plants with clear symptoms of tungro: yellowing and stunting was selected, and then cleared from N. virescens, and then covered with an insect screen. N. virescens collected from the surrounding area were allowed an acquisition period of 5 hours on the selected clump of symptomatic rice plants. After that, two adult N. virescens were taken and immediately brought to each healthy seedling in a sealed glass test tube and allowed an inoculation period of 24 hours. N. virescens were removed from the inoculated seedlings, and each seedling was replanted into a pot (18 cm in diameter). The 10 inoculated and one uninoculated control seedlings treated in Sidrap District were kept in an insect proof greenhouse in Loka Penelitian Penyakit Tungro, Lanrang, Sidrap, Sulawesi Selatan while 10 inoculated and one uninoculated control seedlings treated in Magelang District were kept in an insect proof greenhouse in Faculty of Agriculture, Universitas Gadjah Mada, Daerah Istimewa Yogyakarta. The plants were observed daily for 21 days for the appearance of Tungro symptoms.
2.2. Total RNA Extraction, RT-PCR, and Sequencing
RT-PCR assay was performed to confirm RTSV infection in 20 inoculated rice plants. Total RNA was extracted from leaf samples taken 21 days post inoculation (dpi), using Isogen RNA Extraction Kit (Amersham Pharmacia, UK) following manufacturer instructions except for the use of liquid nitrogen before grinding the samples. The obtained RNA was then used in the cDNA synthesis using oligo d(T) primer and First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) following the standard procedures.
PCR mix was prepared in a volume of 23 μl: 20 μl pureTaq Ready -to-Go PCR Bead (GE Healthcare, USA), 1 μl forward primer (5’-AAACGGTCATTGTGGGGAGGT-3’), 1 μl reverse primer (5’-CAGGCCCAGCAACGACATAA-3’) [10], and 1 μl cDNA sample. The PCR program was as follows Predenaturation: 94 oC for 5 min; 35 cycles of 94 oC for 50 s, 53 oC for 1 min, and 72 oC for 50 s; and Final extention: 72 oC for 5 min.
PCR products were mixed with loading buffer (ThermoFisher Scientific, USA), loaded into 2% agarose gel, and then run in an electrophoresis apparatus at 75 V, 25 mA for 1.5 h. The agarose gel was then stained with 0.1% ethidium bromide for 10 min, and the appearance of the targeted specific band was visualized under a UV transilluminator. Amplicons of randomly selected two RTSV positive samples (one from each sampling location) was purified using ExoSAP-IT reagent (USB Corporation, USA) and then shipped to Integrated Laboratory of Universitas Islam Negeri Sunan Kalijaga, Indonesia to be sequenced bidirectionally using the Sanger method.
2.3. Recombination and Phylogenetic Analyses
On 7 February 2023, the complete genome sequences of six RTSV isolates were downloaded from NCBI GenBank database and aligned using the ClustalW algorithm available in MEGA X v.10.2.4 program [11]. The 5’ and 3’-UTR regions were removed to construct a ‘Full ORFs’ alignment (10,413 bp). Separately, to assess the phylogeny of each ORF, the alignment was also trimmed according to ORF1/Hypothetical protein (1923 bp), ORF2/CP1 (624 bp), ORF3/CP2 (609 bp), ORF4/CP3 (876 bp), ORF5/NTP-binding domain (3537 bp), ORF6/3C-like protease (978 bp), and ORF7/RdRp (1866 bp) sequences, respectively. The complete sequences of ORFS of other isolates in GenBank and complete sequences of CP1 gene of two novel isolates obtained in this study were also included in their respective ORF alignment (Table S1).
Possible recombination events in each alignment were scanned using Recombination Detection Program (RDP) v.5.30 [12] applied different algorithms: RDP, Chimaera, MaxChi, 3Seq, GENECONV, Siscan, and Bootscan, with default parameters. Phylogenetic anomalies supported by fewer than five algorithms with a Bonferroni-corrected P-value < 0.05 were not considered significant.
Maximum Likelihood (ML) phylogenetic trees based on each alignment were built using the best-fit Tamura-Nei-parameter model [13] within MEGA X software, plus uniform rates among sites with 1,000 bootstrap replicates to determine the statistical significance of isolated clusters. The percentage identities between major phylogroups tested at the nucleotide (nt) and amino acid (aa) levels were determined by Sequence Demarcation Tool (SDT) v1.2 freeware [14].
3. RESULTS
3.1. RTSV Transmission
At the end of observation (21 dpi), all 20 inoculated plants showed Tungro-like symptoms such as leaf yellowing and stunted growth with different severity while the two control plants did not show any clear symptoms of viral infection.
3.2. RT-PCR and Sequencing
RT-PCR assay determined that eight inoculated plants, three from Magelang (No. 1, 3, and 7) and five from Sidrap (No. 2, 3, 4, 7, and 9), were tested positive for RTSV while 12 others inoculated and two controls were negative. Since they were assumed from the same inoculum sources, one sample was randomly selected from each location: no. 7 from Magelang and no. 9 from Sindrap are to be sequenced.
3.3. Recombination and Phylogenetic Analyses
RDP5 analysis on the alignment of ‘Full ORFs’ and seven other RTSV genome regions: Hypothetical proteins, CP1, CP2, CP3, NTP-binding domain, 3C-like protease, and RdRp found six significant recombination signals along the full genome sequences of four isolates: Orissa (AM234048), West Bengal (AM234049), Vt6 (AB064963), and AP (KC794785), and a single recombination event on the Phil.1 (U71440) genome sequence was only partially known (Table 1).
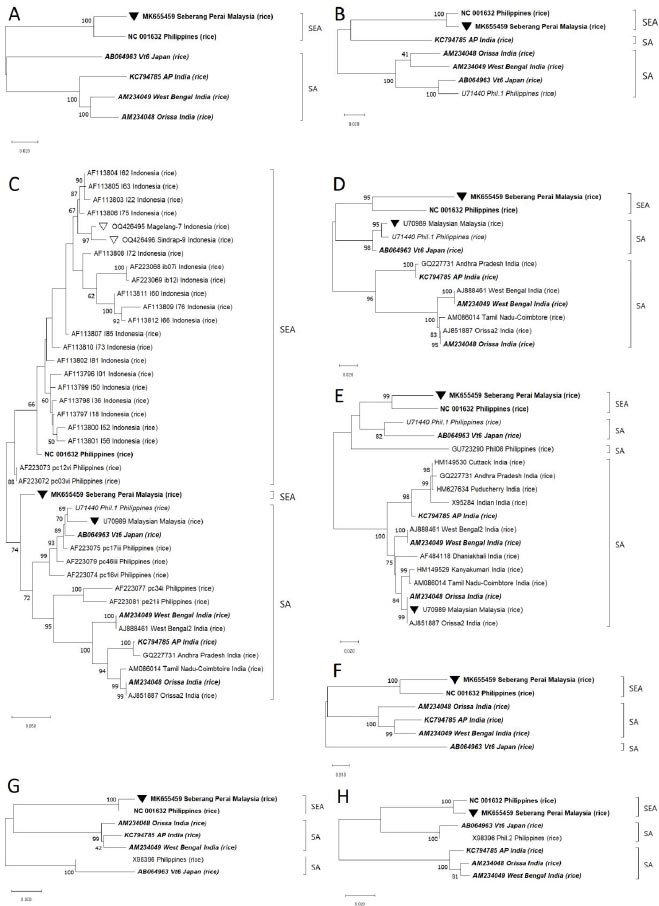
Phylogenetic analysis based on ‘Full ORFs’ deduced that the only available 6 complete isolates belonged to two major lineages that were geographically separated. The Malaysian ‘Seberang Perai’ (MK655459) and Filipinos NC_001632 isolates were positioned in a phylogroup that was proposed to be appropriately named South East Asia (SEA). In the second group, one Japanese ‘Vt6’ (AB064963) was clustered together with three Indian isolates: ‘Orissa’ (AM234048), ‘West Bengal’ (AM234049), and ‘AP’ (KC794785) thus this group was suggested to be called South Asia (SA) (Fig. 2). We then tested the ‘Full ORFs’ phylogrouping against the alignments of seven other RTSV genome regions.
After removal of unclear nt readings, the sequencing analysis recovered 624 bp of nt from each of the two samples, covering the complete CP1 gene. Sequences of the two isolates, named Magelang-7 and Sindrap-9, were registered in NCBI GenBank with accession nos. OQ426495 and OQ426496, respectively. They were then aligned with all other 39 RTSV isolates which complete CP1 gene sequences were registered in GenBank. Phylogenetic analysis based on CP1 gene deduced that the compared 41 isolates were mostly also separated into SEA and SA groups. Magelang-7 and Sindrap-9, together with 19 other Indonesian isolates, formed a subcluster which in turn shared a basal node with three Filipino isolates in the SEA phylogroup. In the second cluster, one Malaysian, one Japanese, and six Filipino isolates were positioned together with seven Indian isolates in the SA phylogroup. Interestingly, MK655459 that was a SEA isolate was shown to be closely related to SA isolates in CP1 tree (Fig. 2C). The 21 Indonesian isolates had 92 – 100% and 94 – 100% identity among them at nt and aa levels, respectively. They also shared 91 – 97% nt and 93 – 100% aa identities with three Filipinos isolates belonging to SEA group, and 85 – 95% nt and 93 – 100% aa identities with 15 isolates in SA group according to SDT matrix analysis.
Further observation showed that two other complete SA isolates: KC794785 and AB064963 were grouped together with SEA isolates for the analysis of different genome regions. Similarly, two partially sequenced isolates: U71440 and ‘Malaysian’ (U70989) that belonged to SA group in CP1 tree were found to be clustered in SEA group in other trees, which indicated the presence of recombination events in the CP1 region of U70989 that were not yet observed by RDP5 software probably due to the lack of known parental isolates (Fig. 2). Therefore, the results of both recombination and phylogenetic analyses confirmed frequent recombination among RTSV isolates.
4. DISCUSSION
This report biologically and molecularly identified RTSV infection in rice cultivated in Sindrap and Magelang Districts. The former location was Sulawesi Island, while the latter is in Central Java, Java Island, with a distance between the islands is about 1,000 km. This showed that RTSV is continuously a major threat in centers of rice cultivation in Indonesia. Although their origins are distantly separated, both Magelang-7 and Sindrap-9 were highly identical at nt and aa levels, at least in the observed CP1 gene. The SDT matrix analysis also demonstrated that Indonesian isolates shared higher identity among themself than with isolates from other countries. The higher nt to aa identity percentages across all comparisons suggested that many nt substitutions were translated into synonymous mutations in the aa sequence of CP1 gene.
The idea that there were two RTSV populations that each had evolved independently in India and the Philippines was put forward [15]. Similarly, the constructed phylogenetic tree based on CP1 gene of this current study hinted that the Philippines could be the diversity center for RTSV since isolates from the country occupied both SEA and SA phylogroups. However, the phylogenetic study involving CP1 gene of two new isolates and 19 old isolates from Bali Island and Subang District, West Java, Java Island [16] suggested that all Indonesian isolates were evolutionary distinct by forming a subcluster separated from three Filipinos isolates in SEA phylogroup (Fig. 2). This was in line with the results of identity percentage analysis by SDT freeware explained in the previous paragraph. Further studies to determine the origin of RTSV will clearly need additional fully sequenced isolates from the Philippines to complement the only complete Filipino isolate (NC_001632) currently in GenBank. Full genome sequence of the divergent Indonesian isolates, which is not yet available, is also essential to understand the evolution of RTSV.
Recombination has been long suspected as the most powerful force in the evolution of RTSV due to the frequent presence of two or three variants of the virus in a single host plant [16] before confirmation by several subsequent researches [15, 17, 18], and the current study which employed the latest bioinformatics software to analyze the old and several newly reported isolates.
Analysis by RDP5 freeware detected recombinant signals in four (AM234048, AM234049, AB064963, and KC794785) out of six complete isolates available in GenBank. Furthermore, phylogenetic analysis by MEGA X freeware positioned ‘Seberang Perai’ isolate (MK655459) in SA group of CP1 tree while clustering the same isolate in SEA group in other trees based on ‘Full ORFs’ and different genome regions, which indicated possible recombination in CP1 gene of MK655459 isolate that was not detected by RDP5 (Fig. 2). The Malaysian MK655459 itself was shown to be a parental donor to two recombinations in the Japanese AB064963 genome (Table 1). Therefore, these results verified that recombination was a common occurrence among RTSV isolates, even those from geographically separated areas, and that currently, NC_001632 is likely the only known recombination-free complete isolate.
| S.No. | Recombinant |
Parents: Major/Minor |
Breakpoints1 (Start/End) |
RDP Implemented Method2 (P-value) |
|---|---|---|---|---|
| 1. | AP (KC794785) (India) | Orissa (AM234048) (India)/Vt6 (AB064963) (Japan) | 129/2042 of ‘Full ORFs’ | R (3.399x10-28) G (8.146x10-18) B (1.058x10-23) M (1.295x10-25) C (1.916x10-23) S (2.211x10-13) 3S (2.789x10-24) |
| 2. | AP (KC794785) (India) | West Bengal (AM234049) (India)/Orissa (AM234048) (India) | 2612/4108 of ‘Full ORFs’ | R (2.731x10-09) G (2.120x10-11) M (2.269x10-13) C (5.871x10-14) S (1.578x10-13) 3S (4.078x10-02) |
| 3. | AP (KC794785) (India) | Vt6 (AB064963)/Orissa (AM234048) (India) | 3401/4077 of ‘Full ORFs’ | R (8.600x10-06) G (4.382x10-09) M (5.598x10-12) C (7.310x10-09) S (4.052x10-18) 3S (4.795x10-03) |
| 3. | West Bengal (AM234049) (India) | Orissa (AM234048) (India)/AP (KC794785) (India) | 1217/2009 of ‘Full ORFs’ | R (4.005x10-08) G (2.683x10-08) M (8.654x10-13) C (8.977x10-08) S (1.506x10-22) 3S (1.593x10-06) |
| 4. | Vt6 (AB064963) (Japan) | NC_001632 (Philippines)/S. Perai (MK655459) (Malaysia) | 2270/2616 of ‘Full ORFs’ | R (2.396x10-10) B (5.303x10-10) M (6.397x10-20) C (2.440x10-12) 3S (4.355x10-10) |
| 5. | Vt6 (AB064963) (Japan) | Orissa (AM234048) (India)/NC_001632 (Philippines) | 8428/9804 of ‘Full ORFs’ | R (4.443x10-04) G (2.655x10-04) M (1.398x10-09) C (4.278x10-11) S (3.866x10-29) 3S (1.094x10-06) |
| 6. | Orissa (AM234048) (India) | West Bengal (AM234049) (India)/AP (KC794785) (India) | 4109/5828 of ‘Full ORFs’ | R (1.623x10-06) G (3.244x10-03) M (3.980x10-08) C (1.609x10-06) S (6.719x10-06) 3S (2.042x10-13) |
| 7. | U71440 (Philippines) | Vt6 (AB064963) (Japan)/S. Perai (MK655459) (Malaysia) | 646/876 of CP3 | R (3.266x10-02) B (2.465x10-02) M (9.864x10-06) C (4.925x10-03) S (7.318x10-10) 3S (5.644x10-06) |
2 R=RDP; G=GENECOV; B=BootScan; M=MaxChi; C=Chimaera; S=Siscan; 3S=3Se.
RTSV can be transmitted without the presence of RTBV by green leafhoppers (N. virescens), but the single RTSV infection was mostly asymptomatic [3]. This study showed rather efficient transmission of Magelang-7 and Sindrap-9 isolates. N. virescens only needed relatively short acquisition and inoculation periods to transmit the virus to healthy plants. However, some of the inoculated plants remained negative for RTSV infection by RT-PCR. This might indicate transmission failure due to inadequate method or some resistance in TN1 rice variety to RTSV which needs additional examination.
CONCLUSION
Diversity analysis of linear ssRNA plant viruses could be the path to their accurate detection and management methods [19]. The data reported here demonstrated that Indonesian isolates are actually unique, and yet their understanding at the molecular level is still very minimal. This could be one of the reasons for the ineffective control of Tungro in the country. Rice varieties expressing RTSV proteins developed in other countries have been shown to be resistant to the virus [20, 21]. Therefore, rigorous population genetics studies of local isolates are clearly required to assemble similar resistant varieties adaptive to the situation in Indonesia.
LIST OF ABBREVIATIONS
| RTBV | = Rice Tungro Bacilliform Virus |
| RTSV | = Rice Tungro Spherical Virus |
| CP 1 | = Coat Protein 1 |
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article is available in The National Center for Biotechnology Information (NCBI) GenBank data base at https://www.ncbi.nlm.nih.gov/nuccore, reference number OQ426495 and OQ426496.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.
SUPPLEMENTARY MATERIALS
Supplementary material is available on the Publisher’s website.