All published articles of this journal are available on ScienceDirect.
Morphological Characterization and Genetic Diversity of Sesame (Sesamum indicum L.) Varieties Cultivated in Ethiopia
Abstract
Introduction:
The experiments were conducted to identify and group sesame genotypes by morphological and genetic characteristics.
Materials and Methods:
Ten nationally released sesame genotypes were obtained from Humera and Melka-Worer research centers and planted at Humera experimental sites. The number of primary branches per plant was found higher (5.93) for M-80. However, the number of nodes per plant was grouped as ‘more’ (24.13) for M-80 compared to Humera-1, which was grouped as the ‘highest’ (26.13). Plant height was tall in all the sesame genotypes studied. Likewise, the number of nodes per plant in all the genotypes was grouped as more nodes per plant i.e. greater than 15. The leaf length ranged from 7.769 cm to 6.67 cm and genotypes, Argene and K-74 had the lowest and the highest values, respectively. The variation was least in qualitative characteristics like flower colour, number of locules per pod, pod pubescence, and type of pod beak. Thousand seed weight and seed yield per kg per ha had a significance level at P=0.05. Pod length was found to be varied among the genotypes and grouped as ‘long “with a mean pod length of 2.44 cm. Hence, genotypes having a long pod length might be preferred for seed production.
The genetic characters of the genotypes were characterized and the level of polymorphism for the selected primers ranged from 56.25% to 100%. ISSR-880 was found with the least level of polymorphism while the other three (ISSR-811, ISSR-812 and ISSR-860,) were found with a high level of polymorphism. Five sesame genotypes (ADI, Humera1, Serkamo, M-80 and E) were clustered and grouped together, while three genotypes, namely, Argena, K-74, and S-Breeder appeared to have their own cluster separately.
Results:
Our study indicates that agro-morphological and genetic characterization of genotypes is useful and effective in determining polymorphisms in the sesame genotypes molecular genotyping, reproducible and it can also produce enough polymorphism to cluster and identify the genotypes.
Conclusion:
The findings presented in this paper may prove useful in the future to include more ISSR markers and induvial genotype investigation for detailed characterization of sesame germplasm that would be important for future breeding and sesame improvement program.
1. INTRODUCTION
Sesame (Sesamum indicum L. 2n=26), is a self-pollinated [1] member of the order Tubiflorae, family Pedaliaceae, and is one of the ancient and most important sources of a quality plant oil used by human beings [2]. It is typically an annual oilseed crop and grown in the tropics and warm subtropics [3]. Sesame seed is rich in nutrition value and due to the quality of oils, this sesame is known as the “Queen of oilseeds” [4]. The lignan compound and its derivatives prevent oxidation of the oil and help it to have long shelf life and stability [5]. Sesame seed contains 50-60% oil, 49.1% fat, 5.3% ash and 21.6% carbohydrates, [6] proteins (18-25%), minerals such as calcium, phosphorus, iron, magnesium, manganese, zinc, and vitamin B1 [7, 8]. For many developing countries, it is an extra rich source of protein [9].
Ethiopia is among the most known and major producers of sesame in sub-Saharan Africa, and Ethiopian sesame is among the highest quality in the world; especially seeds produced in the Humera, northern part of the country are very famous due to their colour, taste and good aroma [10]. One of the oilseeds for which Ethiopia is known in the international market (Fig. 1) is sesame which is a good source of foreign currency after coffee from the agricultural commodities [11]. In the last decade, sesame production and marketing have shown a very significant growth. Between 2011 and 2017, the total area of production (42370 ha, 327,741 tons) and the quantity (293,645 ha, 7,873 hectograms per hectare) of sesame produced grown eightfold [12, 13]. The major contributor of this production achievement is the northwest of Ethiopia, particularly Humera and Gondar [14].
To meet the need for seed certification and to obtain an affordable and maximum yield, we should use high-quality seed, i.e., the seed should be physically as well as genetically pure. To produce quality seeds, different multiplication stages are needed. But a number of factors play a vital role in affecting the quality of the seed, including admixture, cross-pollination, and genetic drift as affected by drought, temperature, edaphic factors, and seed-borne diseases. In order to maintain the required genetic purity standards in the seed fields, field inspection, seed, and seedling inspection and grown out, tests are required.
Farmers and seed producers need an assurance that they are being supplied with quality seed material to know the characteristic of a specific cultivar and be assured of its quality. Thus, there is a need to investigate for quick and reliable methods of cultivar identification and for seed purity testing. The production and distribution of quality seeds of improved varieties can alone result in 20-25 percent increased yield in self-pollinated crops and it is significantly higher cross-pollinated crops. According to the central statistical authority report of 2017/18, the sesame production in Ethiopia was 2,559,039.30 Quintal with a yield of 6.91Q/ha [15]. In the Tigray region, the yield of sesame is almost similar to the national yield i.e., 6.10 Q/t and the area under sesame cultivation is 746,142.65 ha [16]. The most suitable area for sesame production in Ethiopia is low and mild altitude between 500-1300 m above the sea level [17]. However, the productivity per unit area of the available nationally released sesame genotypes is lower compared with other producing countries. The low productivity of sesame (0.72 t/ha) in Ethiopia, particularly in Tigray is often attributed to a lack of certified quality seed of improved varieties. To produce quality seed, improved genetic purity is the primary criteria to yield the inherent potential of improved genotypes. It is not questionable that one factor constraining quality seed production in Ethiopia, particularly in Humera, is a lack of characterized sesame varieties [11].
Seedling and plant are the most widely used methods to identify varietal characteristics based on morphological features. According to the International Union for Protection of New Plant Varieties (UPOV), any new characteristic used in varietal characterization should be clearly defined, accepted and should have a standard method of observation, least or not affected by the environment, accessible to breeders, associated with reasonable costs and efforts. By conducting a growth out test and for varietal identification through morphological characters, the plant and seed characters are well studied and thoroughly documented. The National Test Guidelines need to be developed for the conduct of DUS testing. Such characterization studies are lacking in Ethiopian sesame genotypes.
The most interesting fact is the morphological characteristics that we currently used are not included in all these criteria. Besides, varietal characterization using morphological characterization possesses several unwanted characters such as large space requirement, seasonal dependence, time consumption, tedious and environmental influence. The possible ways to avoid these limitations and to speed up the testing procedures is to use chemical tests or electrophoresis, in addition to morphological markers. The use of DNA profiling techniques will help to establish the varietal identity, to measure the genetic relationship and to assess genetic diversity. Genetic diversity in sesame has been detected by universal markers such as Amplified Fragment Length Polymorphism (AFLP) [18, 19] Sequence-Related Amplified Polymorphisms (SRAP) [20], Random Amplified Polymorphic DNA (RAPD) [21, 22], and inter-simple sequence repeat (ISSR) [23, 24].
It is necessary to use reliable methods to identify and group sesame genotypes. The most preferred and reliable method is the use of DNA fingerprinting to evaluate the genetic purity and to assess the genetic variability in different crops [25-30] The research on cultivar identification in sesame is limited and only seed morphological characteristics cannot distinguish the closely related cultivars. Therefore, more emphasis is needed onvarietal identification of sesame. Hence, the current study was designed with the following objectives:1) identification and grouping of varieties through morphological characters and 2) Identification and grouping of varieties through molecular markers.
2. MATERIALS AND METHODS
2.1. Location of the Experimental Site
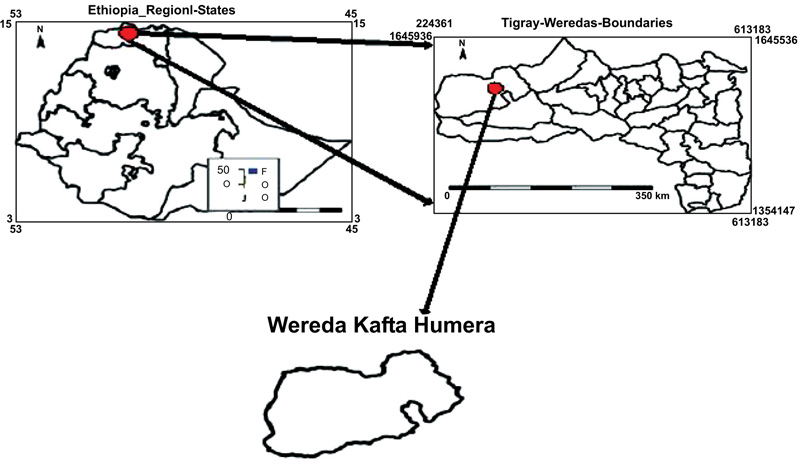
Laboratory and field experiments were performed to characterize the agro-morphological and genetic diversity of sesame genotypes cultivated in Ethiopia. The field experiment was conducted in the Humera agricultural research center, northern Ethiopia, during the rainy season of 2017/2018. Rainy season in Humara starts in late May and ends in late September. Humera research center is located in northern and southern boundaries coincide with 14o15’ to 14027’N latitudes, and 36o 37’ to 37o32’ E longitudes with an altitude of 609 m above the sea level with an annual rainfall of 563.2 mm and min-maximum temperature ranges 18.8-37.6oC (Fig. 2). The dominant soil type of the site is chromic black vertisol and characterized by deep (150 cm) clay textured with 35.66% clay content, silt 35.66%, sandy 38.66% and CEC ranged from 37 to 77 meq/100g of soil [11, 31]. The laboratory experiment was conducted in the Addis Ababa University Department of Biological Sciences, Addis Ababa, Ethiopia.
2.2. Plant Materials and Experimental Design
Ten nationally released sesame genotypes, namely, Setit-1, Hemera-1, Adi, Kelafo 74, Serkamo, E, S, Mehado-80, Arjene and Tate were obtained from Worer agricultural research center and Tigray regional Agricultural research institute and planted in the experimental field site of Humera agricultural research center. The field experiment was laid out in a randomized block design with four replications in a gross plot size of 2.5 X 4.0 m.
2.2.1. Cultural Operations
The land was plowed two times and leveled with the help of plank. The recommended dose of NPK kg per hectare was supplied in the form of urea, single super phosphate/DAP. Fifty percent of nitrogen (20kg per hectare) and the entire quantity of phosphorus and potash were used in the rows 5 centimeters away from the seed row. Remaining fifty percent of N2 was applied 30 days post sowing as a top dressing. For the control of seed-borne diseases, the seeds were treated with thiram at the rate of 3g/kg before sowing. All agronomic practices were applied when needed. Supplementary irrigation was used to maintain soil moisture all the way through the plant growth period. Necessary plant protection measures were taken to control insect pests and diseases.
2.3. Collection of Experimental Data
2.3.1. Sampling Procedure
Data from the qualitative characters were recorded based on visual observation whereas the data of the quantitative characteristics were obtained on a plot basis and measured from representative plants and the average result was obtained . Seven to ten plants were randomly selected from each category: genotype and replication and labeled.
2.3.2. Data Collected
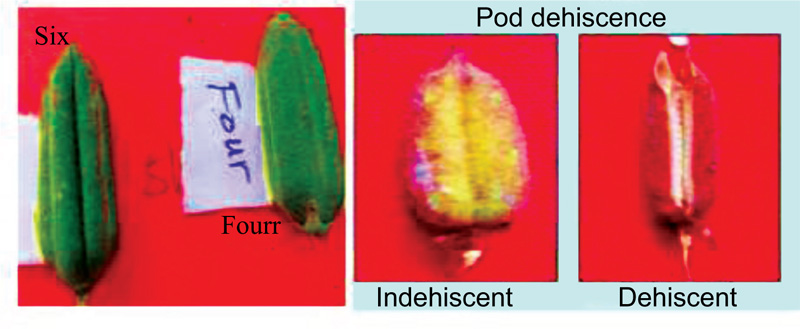
The morphometric data collected on twenty-three characteristics, namely plant height (cm), number of primary branches per plant, number of nodes per plant, internodal length (cm), stem pigmentation, number of leaves per plant, leaf length (cm), leaf shape, leaf color, leaf petiole pigmentation, days to 50 percent flowering, flower petal color, flower hairiness, number of pods per axil, pod length (cm), pod shape, pod beak, pod pubescence, number of locules per pod, pod dehiscence, days to maturity and thousand seed weight (g) were observed, and measured depending on each parameter.
2.4. Genomic DNA Extraction and PCR Amplification
2.4.1. DNA Extraction
DNA extraction was carried out from young leaves of 10 Ethiopian sesame verities following the cetyltrimethylammonium bromide (CTAB) method as described earlier in a study [32]. The purity and integrity of DNA samples were assessed on the basis of an absorbance ratio of 1.80–1.90 at 260/280 using a Nanodrop spectrophotometer (Thermo scientific Nanodrop®, Gene company, China). The concentrations of DNA extracts were quantified using a Nanodrop spectrophotometer and were stored at −20°C.
2.4.2. Primer Screening
A total of 30 SSR primers previously reported [33] were used to test the 10 sesame genotypes. Eleven primers were used to determine the genetic diversity of the genotypes. Polymer Chine Reaction (PCR) was performed according to the annealing temperature of each primer [34].
2.4.3. PCR Amplification and Agarose Gel Electrophoresis
DNA amplification was performed as described in a study [33]. Agarose gel PCR products were separated in 8% non-denaturing polyacrylamide gels (Acr:Bis = 19:1 or 29:1) on a constant voltage of 180 V for 2 ~ 3 h, visualized by silver staining [34].
2.4.4. Statistical Analysis and Data Scoring
The quantitative characteristics for morphological data were analysed using JMP 7.0 statistical program (SAS) Institute, Cary, NC). The mean values of the genotypes in each replication were used for the analysis of variance. Critical differences were calculated at a five percent level of probability (p>0.05).
2.5. Analysis of Amplification Profile
The following parameters were generated using POPGENE 1.32 to describe genetic variation: the percentage of polymorphic loci (P%, 5% criterion) [35], Nei’s genetic diversity (h), Shannon’s information index (i), the observed number of alleles (Na), and the effective number of alleles (Ne) as indicated elsewhere.
To identify genetic variation or diversity patterns among the genotypes, they were represented in the form of a dendrogram that was generated by subjecting the genetic similarity matrix to Unweighted Pair-group Method Arithmetic average (UPGMA) cluster analysis with software NTSYS-pc, Version 2.0 PopGen32, Past.exe, Free Tree (UPGMA), Statistical V5.5, and Arlequin 301 [36].
3. RESULTS
3.1. Identification and Grouping of Sesame Genotypes with Respect to Agro-morphological Characters
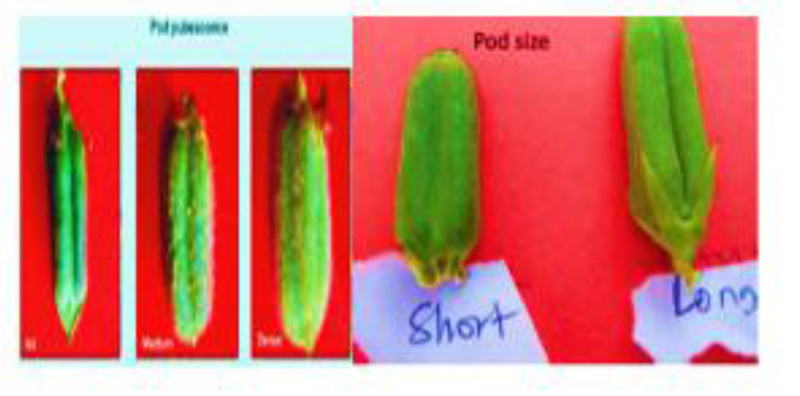
In this study, we evaluated qualitative and quantitative characteristics (plant height, number of primary branches per plant and number of nodes per plant, leaf color, leaf length, leaf petiole, stem pigmentation, number of leaves per plant, leaf length, inter-nod length and number of locules per pod, pod shape, number of pods per plant, pod length, pod beak, pod pubescence Fig. (3), pod dehiscent, yield and 1000seed weight, flower petal color, flower hairiness and day to maturity) and genetic diversity of ten sesame genotypes cultivated in Ethiopia. Based on the results, it is possible to identify and classify genotypes from each other (Tables 1-4).
ANOVA showed significant differences in plant height, number of nodes per plant, leaf length, and internodes length. Two colours (dark green and light green) of seeds were observed. Among the ten genotypes studied, three were light green and seven were dark green. All the ten genotypes evaluated were grouped as a shortleaf length while the other were grouped as long internodes length (Tables 2 and 3). Leaf length differed from 7.769 cm to 6.67 cm and genotypes Argene and K-74 had the lowest and the highest values (Table 2). The analysis of variance revealed a significant difference among the genotypes for leaf length, internodes length, pod length, 1000 seed weight and yield (Qt/ha), indicating the existence of variability among the genotypes for the character studied (Tables 3 and 4). These genotypes were grouped into three groups as light four (Argene, E, S, and K-74), medium four (Humera-1, Serkamo, M-80, and Tate) and dark two (Adi and Setit-1) pigmented based on the intensity of anthocyanin pigmentation.
Genotypes having a greater number of flowers per leaf axil are preferred for breeding programs. Likewise, all of the sesame genotypes in this study were found bearing four flowers per axil. Because the number of capsules per plant is the most contributing parameter for seed yield of sesame plants with three flowers per leaf axil, it is an important resource for breeding programs. The number of pods varied significantly among the different sesamum cultivars (Table 3). The highest number of pods was recorded in the genotype Setit-1 (71.20) and the lowest in Serkamo (43.67). The analysis of variance for pod length, 1000 seed weight and seed yield per Quintal per ha had a significance level at P=0.05 (Table 3). The pod length varied among the genotypes Fig. (4). Based on the variation in pod length, the genotypes were grouped as short (<2.0 cm) and long (>2.0 cm). The pod length in all the genotypes in the current study was observed as long with a mean pod length of 2.44 cm. There were no variations in pod length among the tested genotypes (Table 3). The 1000-seed weight varied significantly with the genotypes. The 1000-seed weight ranged from 2.90 g to 3.43 g with S and Humera-1, ranking the lowest and the highest with a mean weight of 3.11 g, respectively (Table 3) (Fig. 5).
| Genotypes | Plant Height (cm) | Groups | Number of Primary Branches Per Plant | Groups | Number of Nodes Per Plant | Groups |
|---|---|---|---|---|---|---|
| Argene | 136.40 | Tall | 5.47 | High | 22.67 | More |
| E | 150.93 | Tall | 4.93 | High | 23.33 | More |
| S | 146.40 | Tall | 4.53 | High | 21.47 | More |
| TATE | 133.66 | Tall | 4.53 | High | 22.00 | More |
| M-80 | 149.90 | Tall | 5.93 | High | 24.13 | More |
| SERKAMO | 149.90 | Tall | 5.07 | High | 20.40 | More |
| SETIT_1 | 140.40 | Tall | 4.53 | High | 24.00 | More |
| ADI | 128.53 | Tall | 3.27 | High | 21.47 | More |
| `K-74 | 133.47 | Tall | 4.33 | High | 21.20 | More |
| HUMERA-1 | 145.33 | Tall | 3.40 | High | 26.13 | More |
| Mean | 140.49 | 4.60 | 22.68 | |||
| LSD (5%) | ** | NS | ** | |||
| CV (%) | 7.82 | 32 | 15.59 |
| Genotypes | Leaf Colour | Leaf Petiole Pigmentation | Stem Pigmentation | Number of Leaves Per Plant | Groups | Leaf Length (Cm) | Groups | Internode Length (Cm) | Groups | Number of Locules Per Pod |
|---|---|---|---|---|---|---|---|---|---|---|
| Argene | Dark green | Light | Medium | 112.93 | Few | 6.67 | Short | 12.40 | Long | 4 |
| E | Dark green | Light | Medium | 128.67 | Few | 7.28 | Short | 13.20 | Long | 4 |
| S | Dark green | Light | Medium | 102.00 | Few | 7.47 | Short | 13.80 | Long | 4 |
| TATE | Dark green | Medium | Strong | 90.73 | Few | 7.53 | Short | 13.33 | Long | 4 |
| M-80 | Dark green | Medium | Medium | 127.80 | Medium | 7.06 | Short | 11.53 | Long | 4 |
| Serkamo | Light green | Medium | Strong | 102.67 | Few | 7.16 | Short | 12.33 | Long | 4 |
| Setit_1 | Dark green | Dark | Medium | 97.20 | Medium | 7.55 | Short | 11.53 | Long | 4 |
| ADI | Dark green | Dark | Strong | 90.00 | Medium | 7.54 | Short | 10.33 | Long | 4 |
| `K-74 | Light green | Light | Medium | 83.47 | Few | 7.69 | Short | 11.67 | Long | 4 |
| Humera-1 | Light green | Medium | Medium | 98.27 | Few | 7.45 | Short | 11.40 | Long | 4 |
| Mean | 103.37 | 7.34 | 12.15 | 4 | ||||||
| LSD (5%) | NS | * | ** | NS | ||||||
| CV (%) | 40.01 | 8.79 | 15.95 | 0.00 |
Medium: 60-80 Long leaf length: > 10.0 cm long: > 4.0 cm
Many: > 80 Medium leaf length: 8.0-10.0cm [37]
| Genotypes | Pod shape | Number of pods per plant |
Pod length (cm) | Groups | Pod beak | Pod pubescence | Pod Dehiscent |
1000 seed weight (g) |
Groups | Yield (kg/ha) |
|---|---|---|---|---|---|---|---|---|---|---|
| Argene | Oblong | 52.73 | 2.39 | Long | Long beaked | Medium | Indehiscent | 3.00 | Medium | 1959.88 |
| E | Oblong | 57.67 | 2.35 | Long | Long beaked | Medium | Indehiscent | 3.10 | Medium | 2127.57 |
| S | Oblong | 51.00 | 2.33 | Long | Long beaked | Medium | Indehiscent | 3.43 | Medium | 2481.48 |
| TATE | Oblong | 57.67 | 2.45 | Long | Long beaked | Medium | Dehiscent | 2.63 | Medium | 2242.80 |
| M-80 | Oblong | 63.80 | 2.31 | Long | Long beaked | Medium | Indehiscent | 3.33 | Medium | 2377.78 |
| Serkamo | Oblong | 43.67 | 2.37 | Long | Long beaked | Medium | Indehiscent | 3.23 | Medium | 2263.36 |
| SETIT_1 | Oblong | 71.20 | 2.42 | Long | Long beaked | Medium | dehiscent | 3.23 | Medium | 2849.79 |
| ADI | Oblong | 60.53 | 2.64 | Long | Long beaked | Medium | Indehiscent | 3.07 | Medium | 2651.23 |
| `K-74 | Oblong | 58.07 | 2.59 | Long | Long beaked | Medium | Indehiscent | 3.17 | Medium | 3432.09 |
| Humera1 | Oblong | 57.73 | 2.59 | Long | Long beaked | Medium | Dehiscent | 2.90 | Medium | 2194.44 |
| Mean | 57.42 | 2.44 | 3.11 | 2458.04 | ||||||
| `LSD 5% | Ns | *** | *** | ** | ||||||
| CV % | 29.83 | 5.09 | 7.82 | 23.50 |
| Genotypes | Flower Hairiness | Flower Petal Color | Day to Maturity | Groups |
|---|---|---|---|---|
| Argene | Medium | Medium pink | 100 | Late |
| E | Dense | Light pink | 115 | Late |
| S | medium | Medium pink | 115 | Late |
| TATE | NA | NA | 115 | Late |
| M-80 | Medium | Light pink | 115 | Late |
| SERKAMO | Dense | Dark pink | 100 | Late |
| SETIT_1 | Low | Light pink | 95 | Late |
| ADI | NA | NA | 90 | Late |
| K-74 | Medium | Dark pink | 115 | Late |
| Humera -1 | Medium | Medium | 85 | Early |
| `Mean | 93.00 | |||
| SLD (5%) | Ns | |||
| CV | 4.3 |
3.2. Identification and Grouping of Sesame Genotypes Through Electrophoresis
3.2.1. Banding Pattern and ISSR Primers
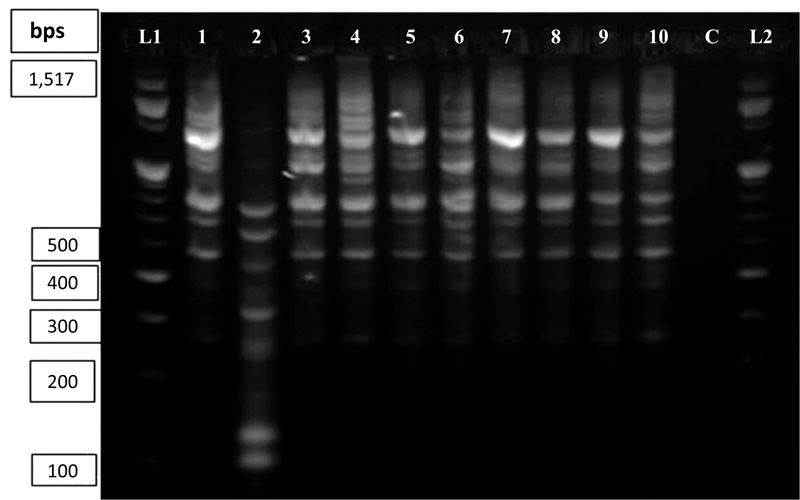
The DNA amplification pattern using ISSR primers in this study was reproducible across gels based on the results from 10 DNA samples. Out of the 30 primers tested initially, 11 of them gave relatively clear banding patterns. NTSYSpc software analysis was used to determine the banding score, percent polymorphism and the number of polymorphic loci for each primer. The band scoring revealed that out of the 11 primers selected, the least scorable bands (7 and 8) were produced by primers 818 and 860, respectively. While the highest banding patterns (19 and 21) were produced by primers 834 and 881, respectively Fig. (6).
Lanes L1 and L2 contain 100bp DNA ladder; Lane C represents the standard check; Lanes 1 to 10 contain amplified sesame products (1= ADI, 2=TATE, 3=HUMERA-1, 4=SERKAMO, 5=E, 6=SETIT-1, 7=ARGENA, 8=K-74, 9=S-BREEDER and 10=M-80).
3.3. Polymorphism and Genetic Diversity
Among the 10 genotypes analysed, POPGENE analysis revealed 376 amplified bands from the 11 selected primers with the level of polymorphism ranging from 56.25% to 100% (Table 5). The number of polymorphic loci was 9 (56.25%) for primer ISSR-880 (least level of polymorphism) and 100% for primers ISSR-811, ISSR-812 and ISSR-860 with numbers of polymorphic loci 15, 10 and 8, respectively. However, the minimum number of polymorphic loci, as revealed by primer ISSR-818, was 4 with 57.14% polymorphism. According to our previous study, the molecular weight across the amplified fragments ranged from 100bp to 1,500bp long.
| Locus | Number of Polymorphic Loci |
Percentage of Polymorphic loci |
na* | ne* | h* | I* |
|---|---|---|---|---|---|---|
| ISSR-811 | 15 | 100% | 2.00 | 1.61 | 0.38 | 0.57 |
| ISSR-812 | 10 | 100% | 2.00 | 1.11 | 0.11 | 0.20 |
| ISSR-818 | 4 | 57.14% | 1.57 | 1.71 | 0.41 | 0.60 |
| ISSR-826 | 8 | 72.73% | 1.73 | 1.54 | 0.11 | 0.22 |
| ISSR-834 | 18 | 94.74% | 1.95 | 1.87 | 0.35 | 0.53 |
| ISSR-848 | 12 | 92.31% | 1.92 | 1.98 | 0.46 | 0.66 |
| ISSR-854 | 9 | 90.00% | 1.90 | 1.54 | 0.49 | 0.69 |
| ISSR-860 | 8 | 100% | 2.00 | 1.11 | 0.35 | 0.53 |
| ISSR-873 | 10 | 90.91% | 1.91 | 1.43 | 0.29 | 0.20 |
| ISSR-880 | 9 | 56.25% | 1.56 | 1.98 | 0.49 | 0.48 |
| ISSR-881 | 17 | 80.95% | 2.00 | 1.38 | 0.27 | 0.69 |
| mean | 10 | - | 2.00 | 1.46 | 0.29 | 0.45 |
| St. dev. | 0.00 | 0.3164 | 0.32 | 0.18 |
*h= Nei’s gene diversity
*I= Shannon’s information index
The number of polymorphic loci is: 11
The percentage of polymorphic loci ranges from 56.25% to 100.00%

3.4. Clustering Analysis
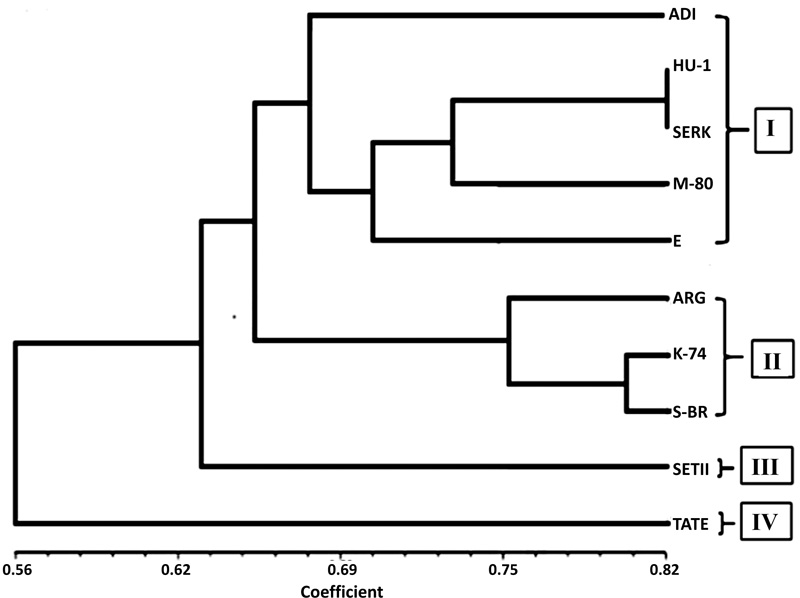
UPGMA analyses were used to construct a dendrogram for the 10 sesame cultivars based on the 376 bands obtained with all the 11 dinucleotide primers, accordingly. In the dendrogram, the sesame varieties named ADI, Humera1, Serkamo, M-80 and E were grouped together in one cluster while three varieties namely, Argena, K-74, and S Breeder were observed to have their own cluster separately (Fig. 7). However, the rest of the cultivars escaped from the major cluster in UPGMA dendrograms.
4. DISCUSSION
Among the ten sesame genotypes studied, the number of primary branches per plant was categorized as ‘more’ (5.93) and more (24.13) number of nodes per plant were recorded for M-80 while the highest (26.13) number of nodes per plant was scored from Humera-1. Plant height was found as ‘tall’ in all the sesame genotypes studied. Likewise, the number of nodes per plant in all the cultivars was classified as more nodes per plant i.e. greater than 15. The number of nodes per plant ranged from 26.13 to 21.47 with the genotype Humera-1 and breeder seed (S), ranking the highest and the lowest, respectively. This could be due to its adaptation to the environmental condition of the experimental area. Similar results were recorded in another study [38].
The minimum and maximum plant height belonged to Adi (128.53 cm) and E (150.93 cm), respectively. Plant height is a useful agronomic character that can help select and group sesame cultivars in ‘short’, ‘medium’ and ‘tall’ categories. The high number of three varieties, namely M-80, Arjene and Serkamo were found with a high number of branches; this may indicate their ability to bear more pod, which may in turn contribute to yield. As a result of this it can be said that selection of plants having more branches is preferred. Variation among sesame genotypes in the number of branches has been reported in another study [39, 40].
Sesame genotypes with a high number of leaves per plant have good yield potential as the number of leaves increases their ability to synthesize nutrients. Several genotypes, such as E, M-80, Argene and breeder seed (S) were found with a high number of leaves and branches. These results revealed that the varieties had good potential to be included in future sesame breeding programs aimed at improving the yield potential of sesame in Humera, northern Ethiopia. Agro-orphological characters such as plant height, number of leaves, and number of pods/plants are not affected by genetic influence [41]. The variation was least for qualitative characters like leaf length, flower colour, number of locules per pod, pod pubescence, and type of pod beak. There was only one flower colour i.e. white with purple shading. The inter-nodal length of the main stem influences the plant height and the number of branches [2]. Analysis of variance showed a significant difference with respect to internodal length with a mean of 12.15 and all the genotypes were grouped as long (long:> 4.0 cm). Similar findings in these characters were earlier reported [42] in castor varieties and hybrids. The variation is genetically controlled and further influenced by environmental conditions and agronomic practices.
The stem pigmentation is among the most important parameters in genotype identification. Based on the stem pigmentation, the varieties were classified as medium (seven varieties) and strongly (three varieties) pigmented. Similar results on stem pigmentation were previously reported [43, 44]. Stem pigmentation can be affected by environmental factors such as temperature and light intensity and it is also genetically controlled [45]. The leaf petiole pigmentation at the peak flowering stage varied among the ten sesamum genotypes. Similar results were also reported by Jain et al. [43], in mung bean varieties. Rajendra et al. [37], grouped sunflower genotypes as present or absent based on their leaf petiole pigmentation. The variation in leaf petiole pigmentation among the genotypes may be due to varied intensity of anthocyanin pigmentation, which is governed genetically and also influenced by the nutritional status of the soil under which the sesame plant is grown.
For arid areas like Humera, early maturing genotypes are more preferred than late matured ones. The genotypes Tate, E, S, M-80, and K-74 had the maximum values for days to flowering (50%) while Humera-1 was found with less value i.e, it takes less time in flowering. Based on flower petal, colour varieties were grouped as light pink (four varieties), medium pink (two varieties) and dark pink (two varieties). The genotypes were further divided into three groups based on the flower hairiness. Among the genotypes, one genotype was low, five genotypes were medium and two genotypes were dense pubescence types. Hairiness is an important morphological character of sesame and found in different parts of the plant [6].
Capsule hairiness was found varied among the genotypes. Two genotypes, namely, Tate and Adi were found with no hairs on stem, leaf, and capsule. Strong or profuse hairs were found in Serkamo and E breeding seed genotypes. However, Setit-1 collected from Humera research institute, exhibited a small stem, less leaves and capsule hairiness. Such characters are an important physical barrier prevention of insect pests and sesame diseases. Haljak et al. [44], studied the hairiness of auricles of flag leaf, flag leaf wax of sheath, ear waxiness, ear density, and peduncle length of nine winter wheat genotypes and suggested that these morphological characters are best for the distinctness of the varieties.
Based on pod number, the varieties were classified into three groups as less (< 40 pods per plant), moderate (40-60 pod per plant) and more (> 60 pods per plant). Among the ten genotypes, seven had moderate (Argene, E, S, Serkamo, K-74, and Humera-1) and three had more (Setit-1, Adi, and M-80) pod numbers per plant. Similar results were observed in chickpea genotypes [46]. The variation in pod number may be due to the pod bearing ability of the genotype and nutrient content of the soil to some extent.
The pod dehiscence varied among the genotypes (Table 3). Genotypes such as Tata, Setit-1 and Humera-1 are grouped as dehiscent and six genotypes, namely, Argene, E, S, M-80, K-74, Adi and Serkamo are grouped as indehiscent types. Likewise, similar results have been reported from castor [42] and jute [47] varieties. The pod dehiscence might be influenced by temperature, especially at the time of flowering. Thousand seed weight is considered as a quality requirement for seed export in the international market. In this study, all the genotypes were grouped as a medium in 1000-seed weight (3.5 g) which implies that all the tested varieties in the current study fulfilled the criteria for export standard i.e. a seed weight of greater than 3g, oil content of 40-50%, and white seed colour are the required criteria.
The average mean for seed yield was determined and found as 2458.04 kg per ha. However, a maximum sesame seed yield of 799.9 kg/ha was reported from Humera, northern Ethiopia [48]. This yield variation could be due to the cultivation conditions, the genetic potential of the genotypes and environment variability. The environment x genotype interaction influences the yield performance of genotypes; as a result, genotypes respond differently to environmental changes in which they are grown [49, 50].
To eliminate the influence of environmental factors on the morphological characters, we analysed the sesame genotypes using ISSR primers and the level of polymorphism among the 10 varieties ranged from 56.25% to 100%. The number of polymorphic loci was 16 for primer 880 (least) and 100% for primers 811, 812 and 860. Bhat et al. [21], evaluated genetic diversity of sesame genotypes and they found a high level of genetic diversity among Indian genotypes. UPGMA analyses were used to construct a dendrogram for the 10 sesame cultivars based on the 376 obtained bands with all the 11 dinucleotide primers. Accordingly, in the dendrogram, the sesame varieties named ADI, Humera1, Serkamo, M-80 and E were grouped together in one cluster and varieties Argena, K-74, and S Breeder appeared to have their own cluster separately Fig. (7). However, the rest of the cultivars get free from the main cluster in the UPGMA dendrogram.
Abdellatef et al. [49], studied 10 sesame genotypes and found the maximum and 92.3% polymorphism while the minimum numbers of fragments were 60% polymorphism. A study [51] reported that when the variation between genotypes is high, the use of a few primers is sufficient. Our results were similar to the findings of other researchers [52-.54]. Zhang et al., [54] who studied 14 agronomic traits and SSR markers to select better performing sesame landraces from China’s germplasm collection.
CONCLUSION
In our study, the information for genotype identification and grouping of sesame genotypes cultivated in Ethiopia through their morphological characteristics has been confirmed with both agromorphological and molecular markers. This study isreliable, and effective in determining polymorphisms in the molecular genotyping of sesame genotypes, reproducible and it can also produce enough polymorphisms to group and identify the genotypes even though most of the molecular markers found polymorphism. To the best of our knowledge, this is the first study on the characterization of Ethiopian sesame genotypes. The findings presented in this paper may be a useful piece in the future to include more ISSR markers and induvial genotype investigation for detailed characterization of sesame germplasm that would be important for future breeding and sesame improvement program.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors would like to express our gratitude to Tigray Agriculture research institute Humera centre for their kind collaboration and providing us the sesame genotypes and allow us to conduct the field experiment in their centre. and special thanks goes to Afar research institute Melka-Worer centre for their provision of sesame genotypes.